Structural analyses of a hypothetical minimal metabolism - PubMed (original) (raw)
Structural analyses of a hypothetical minimal metabolism
Toni Gabaldón et al. Philos Trans R Soc Lond B Biol Sci. 2007.
Abstract
By integrating data from comparative genomics and large-scale deletion studies, we previously proposed a minimal gene set comprising 206 protein-coding genes. To evaluate the consistency of the metabolism encoded by such a minimal genome, we have carried out a series of computational analyses. Firstly, the topology of the minimal metabolism was compared with that of the reconstructed networks from natural bacterial genomes. Secondly, the robustness of the metabolic network was evaluated by simulated mutagenesis and, finally, the stoichiometric consistency was assessed by automatically deriving the steady-state solutions from the reaction set. The results indicated that the proposed minimal metabolism presents stoichiometric consistency and that it is organized as a complex power-law network with topological parameters falling within the expected range for a natural metabolism of its size. The robustness analyses revealed that most random mutations do not alter the topology of the network significantly, but do cause significant damage by preventing the synthesis of several compounds or compromising the stoichiometric consistency of the metabolism. The implications that these results have on the origins of metabolic complexity and the theoretical design of an artificial minimal cell are discussed.
Figures
Figure 1
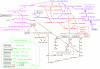
A simplified overview of the metabolic network implemented by a hypothetical minimal genome of 208 protein-coding genes derived by an integrated approach (modified from Gil et al. 2004). Names of substrates freely available for the hypothetical minimal cell are represented inside a frame. Two sink metabolites are labelled in grey. Coenzyme metabolism (except the folate metabolism linked to TTP biosynthesis) is shown in the inset and was not considered in the stoichiometric analysis. Wider arrows in the glycolytic pathway indicate the two steps where ATP is synthesized by substrate-level phosphorylation.
Figure 2
Effect of network size on the deviation of the clustering coefficient from the expected random scenario.
Figure 3
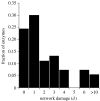
Network damage analysis for the minimal metabolic network. Frequency of deletions causing a given network damage (d), measured as the number of metabolites whose synthesis is prevented by that mutation.
Figure 4
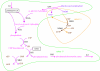
Elementary flux modes (EFM) as a function of the ES. v i Represents denormalized rate of the subset i at every EFM. The input (Ji) and output (Jo) fluxes involved in each ES are also indicated.
Figure 5
The new ES emerging as a consequence of deleting the NDK5 reaction. See table 2 for abbreviations.
Similar articles
- Regularizing capacity of metabolic networks.
Marr C, Müller-Linow M, Hütt MT. Marr C, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2007 Apr;75(4 Pt 1):041917. doi: 10.1103/PhysRevE.75.041917. Epub 2007 Apr 26. Phys Rev E Stat Nonlin Soft Matter Phys. 2007. PMID: 17500931 - Comparing methods for metabolic network analysis and an application to metabolic engineering.
Tomar N, De RK. Tomar N, et al. Gene. 2013 May 25;521(1):1-14. doi: 10.1016/j.gene.2013.03.017. Epub 2013 Mar 26. Gene. 2013. PMID: 23537990 Review. - Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox.
Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. Becker SA, et al. Nat Protoc. 2007;2(3):727-38. doi: 10.1038/nprot.2007.99. Nat Protoc. 2007. PMID: 17406635 - Structural comparison of metabolic networks in selected single cell organisms.
Zhu D, Qin ZS. Zhu D, et al. BMC Bioinformatics. 2005 Jan 14;6:8. doi: 10.1186/1471-2105-6-8. BMC Bioinformatics. 2005. PMID: 15649332 Free PMC article. - The powerful law of the power law and other myths in network biology.
Lima-Mendez G, van Helden J. Lima-Mendez G, et al. Mol Biosyst. 2009 Dec;5(12):1482-93. doi: 10.1039/b908681a. Epub 2009 Oct 2. Mol Biosyst. 2009. PMID: 20023717 Review.
Cited by
- A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola.
Thomas GH, Zucker J, Macdonald SJ, Sorokin A, Goryanin I, Douglas AE. Thomas GH, et al. BMC Syst Biol. 2009 Feb 21;3:24. doi: 10.1186/1752-0509-3-24. BMC Syst Biol. 2009. PMID: 19232131 Free PMC article. - Toward minimal bacterial cells: evolution vs. design.
Moya A, Gil R, Latorre A, Peretó J, Pilar Garcillán-Barcia M, de la Cruz F. Moya A, et al. FEMS Microbiol Rev. 2009 Jan;33(1):225-35. doi: 10.1111/j.1574-6976.2008.00151.x. Epub 2008 Dec 3. FEMS Microbiol Rev. 2009. PMID: 19067748 Free PMC article. Review. - Systems biology perspectives on minimal and simpler cells.
Xavier JC, Patil KR, Rocha I. Xavier JC, et al. Microbiol Mol Biol Rev. 2014 Sep;78(3):487-509. doi: 10.1128/MMBR.00050-13. Microbiol Mol Biol Rev. 2014. PMID: 25184563 Free PMC article. Review. - From essential to persistent genes: a functional approach to constructing synthetic life.
Acevedo-Rocha CG, Fang G, Schmidt M, Ussery DW, Danchin A. Acevedo-Rocha CG, et al. Trends Genet. 2013 May;29(5):273-9. doi: 10.1016/j.tig.2012.11.001. Epub 2012 Dec 6. Trends Genet. 2013. PMID: 23219343 Free PMC article. - Synthetic transitions: towards a new synthesis.
Solé R. Solé R. Philos Trans R Soc Lond B Biol Sci. 2016 Aug 19;371(1701):20150438. doi: 10.1098/rstb.2015.0438. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27431516 Free PMC article. Review.
References
- Albert R, Barabási A.L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97. doi:10.1103/RevModPhys.74.47 - DOI
- Arita M. The metabolic world of Escherichia coli is not small. Proc. Natl Acad. Sci. USA. 2004;101:1543–1547. doi:10.1073/pnas.0306458101 - DOI - PMC - PubMed
- Boeneman K, Crooke E. Chromosomal replication and the cell membrane. Curr. Opin. Microbiol. 2005;8:143–148. doi:10.1016/j.mib.2005.02.006 - DOI - PubMed
- Broder A, Kumar R, Maghoul F, Raghavan P, Rajagopalan S, Stata R, Tomkins A, Wiener J. Graph structure in the web. Comput. Netw. 2000;33:309–320. doi:10.1016/S1389-1286(00)00083-9 - DOI
- de Duve C. Cambridge University Press; Cambridge, UK: 2005. Singularities. Landmarks on the pathways of life.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources