NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development - PubMed (original) (raw)
NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development
Xianfeng Morgan Xu et al. Plant Cell. 2007 May.
Abstract
Vertebrate Tpr and its yeast homologs Mlp1/Mlp2, long coiled-coil proteins of nuclear pore inner basket filaments, are involved in mRNA export, telomere organization, spindle pole assembly, and unspliced RNA retention. We identified Arabidopsis thaliana NUCLEAR PORE ANCHOR (NUA) encoding a 237-kD protein with similarity to Tpr. NUA is located at the inner surface of the nuclear envelope in interphase and in the vicinity of the spindle in prometaphase. Four T-DNA insertion lines were characterized, which comprise an allelic series of increasing severity for several correlating phenotypes, such as early flowering under short days and long days, increased abundance of SUMO conjugates, altered expression of several flowering regulators, and nuclear accumulation of poly(A)+ RNA. nua mutants phenocopy mutants of EARLY IN SHORT DAYS4 (ESD4), an Arabidopsis SUMO protease concentrated at the nuclear periphery. nua esd4 double mutants resemble nua and esd4 single mutants, suggesting that the two proteins act in the same pathway or complex, supported by yeast two-hybrid interaction. Our data indicate that NUA is a component of nuclear pore-associated steps of sumoylation and mRNA export in plants and that defects in these processes affect the signaling events of flowering time regulation and additional developmental processes.
Figures
Figure 1.
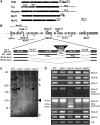
Characterization of NUA T-DNA Insertion Lines. (A) The coiled-coil domain prediction of NUA (At1g79280) using COILS, and comparison with human Tpr, Drosophila Megator, and yeast Mlp1 and Mlp2. Black bars show predicted coiled-coil domains. The dimerization and NPC association domain of Tpr are also depicted. Bar = 200 amino acids (aa). (B) Position of T-DNA insertions and PCR primers. The confirmed insertion sites in nua-1, nua-2, nua-3, and nua-4 are indicated by vertical arrows above and below the schematic exon-intron structure of the NUA gene. Exons are depicted as gray bars and introns as black lines. Horizontal arrows indicate the positions of PCR primers, and brackets indicate the positions of the RT-PCR products shown in (D). The area surrounding the nua-2 insertion site is shown enlarged, and the structure of the ∼200 and ∼300-bp fragments derived from RT-PCR with NUA-D primers in nua-2 is indicated. ex, exon; int, intron. (C) Immunoprecipitation of NUA from Arabidopsis wild-type and nua-1, nua-2, and nua-3 seedlings. Arrowheads depict NUA bands, and asterisks mark unspecific IgG bands also seen without plant extract (data not shown). Molecular weights are indicated on the left. (D) RT-PCR products derived from primer pairs indicated on the right (as shown in [B]) and plant tissues as indicated on the top. Approximate sizes of the fragments amplified with the primer pair for NUA-D in nua-2 are indicated on the left. Two exposures are shown for the fragments amplified with NUA-D primers. The primer combinations in each reaction were as follows: NUA-A, F1 + LP115409; NUA-B, F1 + SALK_057101LP; NUA-C, RP057101.1 + R1(1599); NUA-D, RP069922 + LP069922; NUA-E, CS821281FP + R2(3798); NUA-F, RP079795 + R3(4993). All primer sequences are listed in Supplemental Table 2 online. Tub, tubulin 2 (At5g62690).
Figure 2.
Phenotypic Characteristics of nua Mutant Alleles. (A) Seedlings at 25 d after germination, grown under long-day conditions. (B) Plants at 34 d after germination, grown under long-day conditions. (C) Phyllotaxy defects of nua-1. (D) Silique phenotype of nua-1. (E) Flower phenotype of nua-1.
Figure 3.
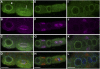
NUA Is Located at the Nuclear Envelope during Interphase and in the Vicinity of the Spindle during Prometaphase. Immunofluorescence images of root tip cell files in interphase ([A] to [D] and [I] to [L]), prometaphase ([A] to [D]), metaphase ([E] to [H]), and late cytokinesis ([I] to [L]). Green, anti-NUA; red in (D), (H), and (L) and magenta in (B), (C), (F), (G), (J), and (K), anti-tubulin; blue in (D), (H), and (L), 4′,6-diamidino-2-phenylindole. (A), (E), and (I) show the green channel only, and (B), (F), and (J) show the red channel only (false colored in magenta). (C), (G), and (K) show the red and green channels, and (D), (H), and (L) show the red, green, and blue channels. The arrowhead in (A) indicates the interphase nuclear envelope, and the arrow in (A) indicates the spindle-like structure in prometaphase. Bars = 10 μm in (D), (H), and (L).
Figure 4.
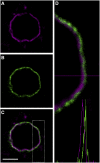
NUA Is Localized at the Inner Side of the Nuclear Envelope, Shown by Colocalization with the Outer Nuclear Envelope/NPC-Localized Protein GFP-WIP1. NUA and GFP-WIP1 were detected by rabbit anti-NUA antibody (A) and mouse anti-GFP antibody (B), respectively, with appropriate secondary antibodies. The overlay image is shown in (C) (green, GFP-WIP1; magenta, NUA). The dashed box in (C) was enlarged, and the green and magenta fluorescence profiles were analyzed in (D). Bar = 5 μm.
Figure 5.
The nua-1 esd4-2 Double Mutant Resembles nua-1 and esd4-2 in Flowering Time and Stunted Growth Characteristics, whereas Additive Effects Exist for Stamen Length. (A) Wild-type, nua-1, esd4-2, and nua-1 esd4-2 seedlings at 21 d after germination, gown under long-day conditions. (B) Plants after 34 d in long-day conditions. (C) Close-up for open flowers. Arrows indicate top of stamens.
Figure 6.
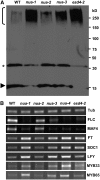
nua Mutant Alleles Lead to Increasing Accumulation of SUMO Conjugates and Altered Expression of Genes Involved in Different Flowering Pathways. (A) Protein extracts from 10-d-old seedlings were probed with an anti-SUMO1 antibody. Free SUMO is reduced (arrowhead), and the amount of SUMO conjugates increased (bracket) in nua mutants, like previously shown for esd4 mutants. The effect increases in severity in the order of nua-2, nua-3, and nua-1. The asterisk indicates the putative SUMO dimer. (B) RT-PCR of tubulin 2 (Tub), FLC, MAF4, FT, SOC1, LFY, MYB33, and MYB65 mRNAs in Arabidopsis wild type, nua-1, nua-2, nua-3, nua-4, and esd4-2.
Figure 7.
The Concentration of ESD4 on the Nuclear Periphery Is Not Abolished in nua Mutants. GFP fluorescence was observed at the nuclear periphery, in root cell files of Arabidopsis seedlings transformed with GFP-ESD4. Bars = 10 μm. (A) Wild-type background. (B) nua-1 background. (C) nua-2 background. (D) nua-3 background.
Figure 8.
Poly(A)+ RNA Accumulates in Nuclei of nua Mutants. Whole-mount in situ hybridization of 10-d-old seedling petioles with fluoresceine-labeled oligo(dT) probe shows increasing severity of nuclear poly(A)+ mRNA retention in nua-2, nua-3, and nua-1. Insets show single nuclei at a higher magnification. All images were taken at identical gain settings. The experiment was repeated twice. Bars = 20 μm.
Similar articles
- NUA Activities at the Plant Nuclear Pore.
Xu XM, Rose A, Meier I. Xu XM, et al. Plant Signal Behav. 2007 Nov;2(6):553-5. doi: 10.4161/psb.2.6.4836. Plant Signal Behav. 2007. PMID: 19704557 Free PMC article. - Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin.
Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, Rothstein SJ, Cui Y. Lu Q, et al. Plant J. 2010 Jan;61(2):259-70. doi: 10.1111/j.1365-313X.2009.04048.x. Epub 2009 Oct 16. Plant J. 2010. PMID: 19843313 - Genetic and environmental changes in SUMO homeostasis lead to nuclear mRNA retention in plants.
Muthuswamy S, Meier I. Muthuswamy S, et al. Planta. 2011 Jan;233(1):201-8. doi: 10.1007/s00425-010-1278-7. Epub 2010 Sep 26. Planta. 2011. PMID: 20872268 - Role of plant RNA-binding proteins in development, stress response and genome organization.
Lorković ZJ. Lorković ZJ. Trends Plant Sci. 2009 Apr;14(4):229-36. doi: 10.1016/j.tplants.2009.01.007. Epub 2009 Mar 13. Trends Plant Sci. 2009. PMID: 19285908 Review. - Diversity of the SUMOylation machinery in plants.
Lois LM. Lois LM. Biochem Soc Trans. 2010 Feb;38(Pt 1):60-4. doi: 10.1042/BST0380060. Biochem Soc Trans. 2010. PMID: 20074036 Review.
Cited by
- Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes.
Elrouby N, Coupland G. Elrouby N, et al. Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17415-20. doi: 10.1073/pnas.1005452107. Epub 2010 Sep 20. Proc Natl Acad Sci U S A. 2010. PMID: 20855607 Free PMC article. - Regulation of cold signaling by sumoylation of ICE1.
Miura K, Hasegawa PM. Miura K, et al. Plant Signal Behav. 2008 Jan;3(1):52-3. doi: 10.4161/psb.3.1.4865. Plant Signal Behav. 2008. PMID: 19704769 Free PMC article. - Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis.
Schweizer N, Ferrás C, Kern DM, Logarinho E, Cheeseman IM, Maiato H. Schweizer N, et al. J Cell Biol. 2013 Dec 23;203(6):883-93. doi: 10.1083/jcb.201309076. J Cell Biol. 2013. PMID: 24344181 Free PMC article. - MdNup62 interactions with MdHSFs involved in flowering and heat-stress tolerance in apple.
Zhang C, An N, Jia P, Zhang W, Liang J, Zhou H, Zhang D, Ma J, Zhao C, Han M, Ren X, Xing L. Zhang C, et al. BMC Plant Biol. 2022 Jul 4;22(1):317. doi: 10.1186/s12870-022-03698-3. BMC Plant Biol. 2022. PMID: 35786201 Free PMC article. - The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex.
Goeres J, Chan PK, Mukhopadhyay D, Zhang H, Raught B, Matunis MJ. Goeres J, et al. Mol Biol Cell. 2011 Dec;22(24):4868-82. doi: 10.1091/mbc.E10-12-0953. Epub 2011 Oct 26. Mol Biol Cell. 2011. PMID: 22031293 Free PMC article.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. - PubMed
- Alcazar-Roman, A.R., Tran, E.J., Guo, S., and Wente, S.R. (2006). Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8 711–716. - PubMed
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
- Chan, G.K., Liu, S.T., and Yen, T.J. (2005). Kinetochore structure and function. Trends Cell Biol. 15 589–598. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases