Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance - PubMed (original) (raw)
. 2007 Jun 28;447(7148):1116-20.
doi: 10.1038/nature05894. Epub 2007 May 21.
Affiliations
- PMID: 17515919
- PMCID: PMC2587297
- DOI: 10.1038/nature05894
Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance
Justin I Odegaard et al. Nature. 2007.
Abstract
Obesity and insulin resistance, the cardinal features of metabolic syndrome, are closely associated with a state of low-grade inflammation. In adipose tissue chronic overnutrition leads to macrophage infiltration, resulting in local inflammation that potentiates insulin resistance. For instance, transgenic expression of Mcp1 (also known as chemokine ligand 2, Ccl2) in adipose tissue increases macrophage infiltration, inflammation and insulin resistance. Conversely, disruption of Mcp1 or its receptor Ccr2 impairs migration of macrophages into adipose tissue, thereby lowering adipose tissue inflammation and improving insulin sensitivity. These findings together suggest a correlation between macrophage content in adipose tissue and insulin resistance. However, resident macrophages in tissues display tremendous heterogeneity in their activities and functions, primarily reflecting their local metabolic and immune microenvironment. While Mcp1 directs recruitment of pro-inflammatory classically activated macrophages to sites of tissue damage, resident macrophages, such as those present in the adipose tissue of lean mice, display the alternatively activated phenotype. Despite their higher capacity to repair tissue, the precise role of alternatively activated macrophages in obesity-induced insulin resistance remains unknown. Using mice with macrophage-specific deletion of the peroxisome proliferator activated receptor-gamma (PPARgamma), we show here that PPARgamma is required for maturation of alternatively activated macrophages. Disruption of PPARgamma in myeloid cells impairs alternative macrophage activation, and predisposes these animals to development of diet-induced obesity, insulin resistance, and glucose intolerance. Furthermore, gene expression profiling revealed that downregulation of oxidative phosphorylation gene expression in skeletal muscle and liver leads to decreased insulin sensitivity in these tissues. Together, our findings suggest that resident alternatively activated macrophages have a beneficial role in regulating nutrient homeostasis and suggest that macrophage polarization towards the alternative state might be a useful strategy for treating type 2 diabetes.
Figures
Figure 1
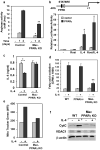
PPARγ regulates alternative macrophage activation. a, Decreased induction of arginase activity by IL-4 in PPARγ null macrophages. BMDM from control and Mac-PPARγ KO mice were stimulated with IL-4 (10ng/ml) for 24 or 48 hours prior to quantification of cell-associated arginase activity. b, Activation of arginase I promoter by PPARγ/RXR heterodimers. c, PPARγ is required for suppression of IL-6 production in alternatively activated macrophages. Macrophages pre-treated with IL-4 (10 ng/ml) for 24 hours were subsequently stimulated with LPS (5 ng/ml) for 6 hours (TNFα) or 24 hours (IL-6). d, PPARγ is required for macrophage oxidative metabolism. Fatty acid oxidation rates were quantified in control, PPARδ null and PPARγ null BMDMs 96 hours after stimulation with IL-4. e-f, IL-4 fails to induce mitochondrial biogenesis in PPARγ deficient macrophages, as measured by (e) Mito Tracker Green and (f) CytC and VDAC1 protein levels. Equivalent loading was confirmed by immunoblotting for β-actin. Rosiglitazone (Rosi).
Figure 2
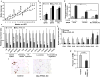
Mac-PPARγ KO mice are less susceptible to infection by Leishmania major. a, Footpad swelling in control and Mac-PPARγ KO mice after infection with L. major (n=5/genotype). b, Decreased necrosis in footpads of Mac-PPARγ KO mice.
Figure 3
Alterations in adipose tissue mass and function in Mac-PPARγ KO mice. a, Weight gain of control and Mac-PPARγ KO mice on a HFD. Arrow denotes period when mice were fasted for glucose and insulin tolerance tests. b, Body composition as determined by DEXA (n=5/genotype). c, Q-PCR analyses of gonadal adipose tissue gene expression. Relative transcript levels of genes involved in adipocyte differentiation and function. Lpl, lipoprotein lipase; Cd36, fatty acid translocase; Slc27a1, fatty acid transporter 1; Slc2a4, glucose transporter 4; Fabp4, fatty acid binding protein 4; Lipe, hormone sensitive lipase; Fasn, fatty acid synthase; Acaca, acetyl-Coenzyme A carboxylase a; Acox1, acyl-Coenzyme A oxidase 1; Cpt1a, carnitine palmitoyltransferase 1a; Acadm and Acadl, medium- and long-chain acyl-CoA dehydrogenase; Adipoq, adiponectin; Srebf1, sterol regulatory element binding factor 1c. d, Co-culture of macrophages with adipocytes decreases insulin-stimulated glucose uptake. BMDMs from control or Mac-PPARγ KO mice were co-cultured with differentiated 3T3-L1 adipocytes for 48 hours. 2-deoxyglucose uptake was assessed 30 minutes after stimulation with insulin. e, Q-PCR analyses of macrophage gene expression in WAT from control and Mac-PPARγ KO mice. Emr1, F4/80; Cd68, macrosialin; Arg1, arginase I; Mrc1, mannose receptor; Clec7a, dectin-1; Retnla, resistin like alpha; Nos2, inducible nitric oxide synthase; IL-6, interleukin-6. f-g, Macrophage content of epididymal adipose tissue. Representative sections of epididymal fad pads stained with F4/80 (Emr1) antibody (f). Fraction of ATMs is equal to F4/80-stained cells/total cells counted in the fields (g), and statistically analyzed using the paired t-test.
Figure 4
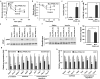
Impaired glucose homeostasis in high fat fed male Mac-PPARγ KO mice. a, Oral glucose tolerance tests (1 g/kg) in male mice after 19 weeks of feeding the HFD (n=5/genotype). b, Insulin tolerance test. Obese mice were fasted for 4 hours prior to intraperitoneal injection of insulin (0.65 u/kg). c, Fasting serum insulin levels in control and Mac-PPARγ KO mice after 4 hr fast. d, Homa-IR index of insulin sensitivity (insulin [ng/ml] x glucose [mM]). e-f, Decreased insulin signaling in obese Mac-PPARγ KO mice. Control and Mac-PPARγ KO mice were injected with insulin (5 mU/g), and cellular lysates were immunoblotted for total and serine phosphorylated (S473) Akt; liver (e) and quadriceps (f). g-h, Relative transcript levels of genes involved in β-oxidation and oxidative phosphorylation, and of transcriptional regulators controlling these pathways in quadriceps (g) and liver (h). i, Circulating levels of adiponectin in control and Mac-PPARγ KO mice.
Similar articles
- Adipogenic miR-27a in adipose tissue upregulates macrophage activation via inhibiting PPARγ of insulin resistance induced by high-fat diet-associated obesity.
Yao F, Yu Y, Feng L, Li J, Zhang M, Lan X, Yan X, Liu Y, Guan F, Zhang M, Chen L. Yao F, et al. Exp Cell Res. 2017 Jun 15;355(2):105-112. doi: 10.1016/j.yexcr.2017.03.060. Epub 2017 Mar 30. Exp Cell Res. 2017. PMID: 28365247 - Loss of PPAR gamma in immune cells impairs the ability of abscisic acid to improve insulin sensitivity by suppressing monocyte chemoattractant protein-1 expression and macrophage infiltration into white adipose tissue.
Guri AJ, Hontecillas R, Ferrer G, Casagran O, Wankhade U, Noble AM, Eizirik DL, Ortis F, Cnop M, Liu D, Si H, Bassaganya-Riera J. Guri AJ, et al. J Nutr Biochem. 2008 Apr;19(4):216-28. doi: 10.1016/j.jnutbio.2007.02.010. Epub 2007 Jul 6. J Nutr Biochem. 2008. PMID: 17618105 - Peroxisome proliferator-activated receptor gamma activation promotes infiltration of alternatively activated macrophages into adipose tissue.
Stienstra R, Duval C, Keshtkar S, van der Laak J, Kersten S, Müller M. Stienstra R, et al. J Biol Chem. 2008 Aug 15;283(33):22620-7. doi: 10.1074/jbc.M710314200. Epub 2008 Jun 9. J Biol Chem. 2008. PMID: 18541527 - Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance.
Lee BC, Lee J. Lee BC, et al. Biochim Biophys Acta. 2014 Mar;1842(3):446-62. doi: 10.1016/j.bbadis.2013.05.017. Epub 2013 May 22. Biochim Biophys Acta. 2014. PMID: 23707515 Free PMC article. Review. - Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity.
Heilbronn LK, Campbell LV. Heilbronn LK, et al. Curr Pharm Des. 2008;14(12):1225-30. doi: 10.2174/138161208784246153. Curr Pharm Des. 2008. PMID: 18473870 Review.
Cited by
- Interferon regulatory factor 4 regulates obesity-induced inflammation through regulation of adipose tissue macrophage polarization.
Eguchi J, Kong X, Tenta M, Wang X, Kang S, Rosen ED. Eguchi J, et al. Diabetes. 2013 Oct;62(10):3394-403. doi: 10.2337/db12-1327. Epub 2013 Jul 8. Diabetes. 2013. PMID: 23835343 Free PMC article. - Molecular understanding and clinical aspects of tumor-associated macrophages in the immunotherapy of renal cell carcinoma.
Liu H, Lv Z, Zhang G, Yan Z, Bai S, Dong D, Wang K. Liu H, et al. J Exp Clin Cancer Res. 2024 Aug 22;43(1):242. doi: 10.1186/s13046-024-03164-y. J Exp Clin Cancer Res. 2024. PMID: 39169402 Free PMC article. Review. - Differential effects of high-fat-diet rich in lard oil or soybean oil on osteopontin expression and inflammation of adipose tissue in diet-induced obese rats.
Wang X, Cheng M, Zhao M, Ge A, Guo F, Zhang M, Yang Y, Liu L, Yang N. Wang X, et al. Eur J Nutr. 2013 Apr;52(3):1181-9. doi: 10.1007/s00394-012-0428-z. Epub 2012 Jul 31. Eur J Nutr. 2013. PMID: 22847642 - Redox control of inflammation in macrophages.
Brüne B, Dehne N, Grossmann N, Jung M, Namgaladze D, Schmid T, von Knethen A, Weigert A. Brüne B, et al. Antioxid Redox Signal. 2013 Aug 20;19(6):595-637. doi: 10.1089/ars.2012.4785. Epub 2013 Mar 6. Antioxid Redox Signal. 2013. PMID: 23311665 Free PMC article. Review. - Navigating Lipodystrophy: Insights from Laminopathies and Beyond.
Krüger P, Hartinger R, Djabali K. Krüger P, et al. Int J Mol Sci. 2024 Jul 23;25(15):8020. doi: 10.3390/ijms25158020. Int J Mol Sci. 2024. PMID: 39125589 Free PMC article. Review.
References
- Hotamisligil GS. Inflammation and metabolic disorders. 2006;444:860–867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 DK062386-05/DK/NIDDK NIH HHS/United States
- R01 HL076746/HL/NHLBI NIH HHS/United States
- R01 DK066525/DK/NIDDK NIH HHS/United States
- R01 HL076746-05/HL/NHLBI NIH HHS/United States
- K08 DK062386-06/DK/NIDDK NIH HHS/United States
- F31 AI066402/AI/NIAID NIH HHS/United States
- AI007290/AI/NIAID NIH HHS/United States
- R01 HL076746-04/HL/NHLBI NIH HHS/United States
- R01 HL076746-03/HL/NHLBI NIH HHS/United States
- R01 DK076760/DK/NIDDK NIH HHS/United States
- T32 AI007290/AI/NIAID NIH HHS/United States
- R01 DK066525-05/DK/NIDDK NIH HHS/United States
- K08 DK062386/DK/NIDDK NIH HHS/United States
- R01 HL076746-01/HL/NHLBI NIH HHS/United States
- R01 HL076746-02/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases