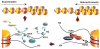Transcription and RNA interference in the formation of heterochromatin - PubMed (original) (raw)
Review
Transcription and RNA interference in the formation of heterochromatin
Shiv I S Grewal et al. Nature. 2007.
Abstract
Transcription in heterochromatin seems to be an oxymoron--surely the 'silenced' form of chromatin should not be transcribed. But there have been frequent reports of low-level transcription in heterochromatic regions, and several hundred genes are found in these regions in Drosophila. Most strikingly, recent investigations implicate RNA interference mechanisms in targeting and maintaining heterochromatin, and these mechanisms are inherently dependent on transcription. Silencing of chromatin might involve trans-acting sources of the crucial small RNAs that carry out RNA interference, but in some cases, transcription of the region to be silenced seems to be required--an apparent contradiction.
Figures
Figure 1
Changes in histone modification implicated in the switch from a euchromatic to a heterochromatic state. Active genes are frequently marked by H3K4me (but see Berger, this issue); this mark must presumably be removed by LSD1 (not yet characterized in Drosophila). H3K9 is normally acetylated in euchromatin, and this mark must be removed by a histone deacetylase, typically HDAC1. Phosphorylation of H3S10 can interfere with methylation of H3K9; dephosphorylation may involved a phosphatase targeted through the carboxyl terminus of the JIL1 kinase. These transitions set the stage for acquisition of the modifications associated with silencing, including methylation of H3K9 by SU(VAR)3-9 or another HMT, binding of HP1, and subsequent methylation of H4K20 by SUV4-20, an enzyme recruited by HP1. Other silencing marks, such as methylation of H3K27 by E(Z) (not shown), appear to be relevant in some domains, although this mark is more prominently used by the Polycomb system. Supporting data comes from genetic identification of modifiers of PEV, biochemical characterization of the activities of such modifiers, and tests of protein-protein interactions . (Adapted from10)
Figure 2
Variegating phenotypes. While alternative chromatin packaging states can be inherited, they do switch at a low frequency, resulting in a variegating phenotype within a clone of cells. Picture: on the left, a fly eye; on the right a sectored colony of yeast S. pombe
Figure 3
HP1 interacts with H3K9me2/3 through its chromo domain, and with SU(VAR)3-9 through its chromoshadow domain. (Adapted from ref .)
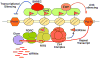
Figure 4
Model showing RNAi-mediated heterochromatin assembly and silencing in S. pombe. Centromeric repeat (dg and dh) transcripts produced by RNA polymerase II are processed by the RNAi machinery, including RITS and RDRC effector complexes that interact with each other and localize across heterochromatic domains. The slicer activity of Ago1 (a component of RITS) and the RNA-dependent RNA polymerase activity of Rdp1 (a subunit of RDRC) are required for processing the repeat transcripts into siRNAs. siRNA-guided cleavage of nascent transcripts by Ago1 might make them preferential substrates for Rdp1 to generate dsRNA, which in turn is processed into siRNAs by Dicer. siRNAs are believed to mediate targeting of histone modifying activities including the Clr4 complex. This process most likely involves siRNA base pairing with nascent transcripts, but the precise mechanism remains undefined. siRNAs produced by heterochromatin-bound RNAi “factories” might also prime assembly of RISC-like complexes capable of mounting a classic RNAi response. Methylation of H3K9 by Clr4 is necessary for stable association of RITS with heterochromatic loci, apparently via binding of the Chp1 chromodomain. This methylation event also recruits Swi6**,** which along with other factors mediates spreading of various effectors such as SHREC. SHREC may facilitate proper positioning of nucleosomes to organize the higher-order chromatin structure essential for diverse heterochromatin functions, including transcriptional gene silencing. Swi6 also recruits an antisilencing protein, Epe1, that modulates heterochromatin to facilitate repeat transcription in addition to other functions. The system as a whole maintains a balance that results in loss of expression from reporter genes inserted within the domain.
Similar articles
- Accumulation of RNA on chromatin disrupts heterochromatic silencing.
Brönner C, Salvi L, Zocco M, Ugolini I, Halic M. Brönner C, et al. Genome Res. 2017 Jul;27(7):1174-1183. doi: 10.1101/gr.216986.116. Epub 2017 Apr 12. Genome Res. 2017. PMID: 28404620 Free PMC article. - RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast.
Creamer KM, Partridge JF. Creamer KM, et al. Wiley Interdiscip Rev RNA. 2011 Sep-Oct;2(5):632-46. doi: 10.1002/wrna.80. Epub 2011 Mar 23. Wiley Interdiscip Rev RNA. 2011. PMID: 21823226 Free PMC article. Review. - Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription.
Jih G, Iglesias N, Currie MA, Bhanu NV, Paulo JA, Gygi SP, Garcia BA, Moazed D. Jih G, et al. Nature. 2017 Jul 27;547(7664):463-467. doi: 10.1038/nature23267. Epub 2017 Jun 22. Nature. 2017. PMID: 28682306 Free PMC article. - The Paf1 complex represses small-RNA-mediated epigenetic gene silencing.
Kowalik KM, Shimada Y, Flury V, Stadler MB, Batki J, Bühler M. Kowalik KM, et al. Nature. 2015 Apr 9;520(7546):248-252. doi: 10.1038/nature14337. Epub 2015 Mar 25. Nature. 2015. PMID: 25807481 Free PMC article. - Slicing and spreading of heterochromatic silencing by RNA interference.
Locke SM, Martienssen RA. Locke SM, et al. Cold Spring Harb Symp Quant Biol. 2006;71:497-503. doi: 10.1101/sqb.2006.71.062. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381332 Review.
Cited by
- The Role of piRNA-Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in Drosophila melanogaster.
Lee YC. Lee YC. PLoS Genet. 2015 Jun 4;11(6):e1005269. doi: 10.1371/journal.pgen.1005269. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26042931 Free PMC article. - Argonaute and the nuclear RNAs: new pathways for RNA-mediated control of gene expression.
Gagnon KT, Corey DR. Gagnon KT, et al. Nucleic Acid Ther. 2012 Feb;22(1):3-16. doi: 10.1089/nat.2011.0330. Epub 2012 Jan 27. Nucleic Acid Ther. 2012. PMID: 22283730 Free PMC article. Review. - Intercepting noncoding messages between germline and soma.
Gao S, Liu Y. Gao S, et al. Genes Dev. 2012 Aug 15;26(16):1774-9. doi: 10.1101/gad.199992.112. Genes Dev. 2012. PMID: 22895251 Free PMC article. - Methylation of lysine 9 in histone H3 directs alternative modes of highly dynamic interaction of heterochromatin protein hHP1β with the nucleosome.
Munari F, Soeroes S, Zenn HM, Schomburg A, Kost N, Schröder S, Klingberg R, Rezaei-Ghaleh N, Stützer A, Gelato KA, Walla PJ, Becker S, Schwarzer D, Zimmermann B, Fischle W, Zweckstetter M. Munari F, et al. J Biol Chem. 2012 Sep 28;287(40):33756-65. doi: 10.1074/jbc.M112.390849. Epub 2012 Jul 19. J Biol Chem. 2012. PMID: 22815475 Free PMC article. - RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy.
Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, van der Maarel SM, Filippova GN, Tapscott SJ. Snider L, et al. Hum Mol Genet. 2009 Jul 1;18(13):2414-30. doi: 10.1093/hmg/ddp180. Epub 2009 Apr 9. Hum Mol Genet. 2009. PMID: 19359275 Free PMC article.
References
- Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 2006;22:330–8. - PubMed
- Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev. 2002;12:178–87. - PubMed
- Sugiyama T, et al. SHREC, an Effector Complex for Heterochromatic Transcriptional Silencing. Cell. 2007;128:491–504. - PubMed
- Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases