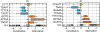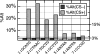A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing - PubMed (original) (raw)
A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing
Rodger B Voelker et al. Genome Res. 2007 Jul.
Abstract
Orthologous mammalian introns contain many highly conserved sequences. Of these sequences, many are likely to represent protein binding sites that are under strong positive selection. In order to identify conserved protein binding sites that are important for splicing, we analyzed the composition of intronic sequences that are conserved between human and six eutherian mammals. We focused on all completely conserved sequences of seven or more nucleotides located in the regions adjacent to splice-junctions. We found that these conserved intronic sequences are enriched in specific motifs, and that many of these motifs are statistically associated with either alternative or constitutive splicing. In validation of our methods, we identified several motifs that are known to play important roles in alternative splicing. In addition, we identified several novel motifs containing GCT that are abundant and are associated with alternative splicing. Furthermore, we demonstrate that, for some of these motifs, conservation is a strong indicator of potential functionality since conserved instances are associated with alternative splicing while nonconserved instances are not. A surprising outcome of this analysis was the identification of a large number of AT-rich motifs that are strongly associated with constitutive splicing. Many of these appear to be novel and may represent conserved intronic splicing enhancers (ISEs). Together these data show that conservation provides important insights into the identification and possible roles of cis-acting intronic sequences important for alternative and constitutive splicing.
Figures
Figure 1.
Example of mammalian genomic alignment showing conserved exonic and intronic sequences. Shown is a small portion of the human gene LMO7 aligned against the orthologous region of the six mammalian genomes used in this study. (A) Four exons (represented as boxes) and their corresponding introns. The central graphic indicates the observed splicing events. Below that, the conservation is represented as a histogram where the height is proportional to the degree of conservation. (B) An expanded view of the sequence flanking the 5′ splice-junction of the second, alternatively spliced, exon. The actual sequence is displayed below the conservation histogram. Boxes are placed around conserved sequences (CSs). The original graphics were drawn using the UCSC Genome Browser (
; Kent et al. 2002).
Figure 2.
Schematic representation of mammalian introns detailing the regions used in this study. The positions of the 5′ and 3′ splice-junctions are indicated as 5′ SJ and 3′ SJ. Sequence logos, composed from 5000 randomly sampled human introns, are used to show the frequency composition of the splice-junctions. The intronic regions that are the basis of this study are indicated as DI (donor intronic) and AI (acceptor intronic).
Figure 3.
Distribution of total lengths of CSs found in donor and acceptor intronic regions and associations between CS length and alternative splicing. (A,B) Distributions of the lengths of the CSs found in the donor or acceptor intronic regions. In cases where more than one CS was found in a particular intron, the lengths were combined (total length). For each data set, the bin width is equivalent to two bases. The number of CSs in each bin is indicated along the _Y_-axis. The inset plots are the same data displayed using a log scaled _Y_-axis to better visualize the longer CSs. (C) The relationship between the percentage of introns that are alternatively spliced and the total length of CS found within the intron. The horizontal bar indicates the average percentage of splice-junctions that are alternatively spliced (3%). (D) The number (and corresponding percentages) of all alternatively spliced (Alt+) or constitutively spliced (Alt−) splice-junctions that contain a CS in the donor (DI) or acceptor (AI) intronic flanking sequence.
Figure 4.
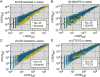
Scatter-plots for the counts of all _n_-mers (4–7 nt) in the CS samples (NCS) vs. the counts in the corresponding random samples (NRS). Overlaid on plots A and C are the _n_-mers that were significantly enriched (according to the confidence intervals described in the Supplemental Materials and Methods) in the donor intronic (DI) and acceptor intronic (AI) regions. Overlaid on plots B and D are all _n_-mers containing the substrings indicated. These substrings are examples of substrings that are enriched in the corresponding regions.
Figure 5.
Samples of GCCS clusters derived from the donor intronic (DI) region. Shown are the graph clusters representing the clustered _n_-mers used to construct the CSMs for the putative Fox and QKI protein binding sites (Fig. 7, DI-1 and DI-2, respectively). Vertices are colored according to their conservation Z-score (see color key). The graphs were drawn using GraphViz (
).
Figure 6.
Box-plots showing the distributions of TA-scores observed for several representative CSMs. The greatest common substrings (GCS) for each CSM are shown to the left. CSMs that were significantly enriched in _n_-mers associated with alternative splicing are shown in red, those significantly associated with constitutive splicing are in blue, and no association is shown in yellow.
Figure 7.
Intronic conserved sequence motifs (CSMs) showing significant associations with alternative (above line) or constitutive splicing (below line) are shown against a schematic representation of an intron to indicate the region within which they are located. Motifs marked with an asterisk are compositionally similar to the equivalently numbered motif in the other region.
Figure 8.
Association between motif conservation and alternative splicing for several 5-mers. Vertical bars represent the percentage of occurrences of each 5-mer that was observed in the DI region of an alternatively spliced intron. The lower horizontal bar indicates the average for all non-CS 5-mers. The upper horizontal bar indicates the average for all CS 5-mers.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.
Love AMA, Edwards C, Cai RY, Gibbs V. Love AMA, et al. Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article. - Repetitive Negative Thinking As a Transdiagnostic Prospective Predictor of Depression and Anxiety Symptoms in Neurodiverse First-Semester College Students.
McKenney EE, Brunwasser SM, Richards JK, Day TC, Kofner B, McDonald RG, Williams ZJ, Gillespie-Lynch K, Kang E, Lerner MD, Gotham KO. McKenney EE, et al. Autism Adulthood. 2023 Dec 1;5(4):374-388. doi: 10.1089/aut.2022.0078. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116057 Free PMC article. - "I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.
Huang Y, Trollor JN, Foley KR, Arnold SRC. Huang Y, et al. Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article. - Pharmacological treatments in panic disorder in adults: a network meta-analysis.
Guaiana G, Meader N, Barbui C, Davies SJ, Furukawa TA, Imai H, Dias S, Caldwell DM, Koesters M, Tajika A, Bighelli I, Pompoli A, Cipriani A, Dawson S, Robertson L. Guaiana G, et al. Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
Cited by
- Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules.
Wang Y, Ma M, Xiao X, Wang Z. Wang Y, et al. Nat Struct Mol Biol. 2012 Oct;19(10):1044-52. doi: 10.1038/nsmb.2377. Epub 2012 Sep 16. Nat Struct Mol Biol. 2012. PMID: 22983564 Free PMC article. - Context-dependent robustness to 5' splice site polymorphisms in human populations.
Lu ZX, Jiang P, Cai JJ, Xing Y. Lu ZX, et al. Hum Mol Genet. 2011 Mar 15;20(6):1084-96. doi: 10.1093/hmg/ddq553. Epub 2010 Dec 28. Hum Mol Genet. 2011. PMID: 21224255 Free PMC article. - Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2.
Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Zhang C, et al. Genes Dev. 2008 Sep 15;22(18):2550-63. doi: 10.1101/gad.1703108. Genes Dev. 2008. PMID: 18794351 Free PMC article. - A systematic analysis of intronic sequences downstream of 5' splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation.
Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, Parkinson J, Frey BJ, Rommens JM, Blencowe BJ. Aznarez I, et al. Genome Res. 2008 Aug;18(8):1247-58. doi: 10.1101/gr.073155.107. Epub 2008 May 2. Genome Res. 2008. PMID: 18456862 Free PMC article. - Intronic motif pairs cooperate across exons to promote pre-mRNA splicing.
Ke S, Chasin LA. Ke S, et al. Genome Biol. 2010;11(8):R84. doi: 10.1186/gb-2010-11-8-r84. Epub 2010 Aug 12. Genome Biol. 2010. PMID: 20704715 Free PMC article.
References
- Agresti A., Coull B.A., Coull B.A. Approximate is better than exact for interval estimation of binomial proportions. Am. Stat. 1998;52:119–126.
- Barreau C., Paillard L., Mereau A., Osborne H.B., Paillard L., Mereau A., Osborne H.B., Mereau A., Osborne H.B., Osborne H.B. Mammalian CELF/Bruno-like RNA-binding proteins: Molecular characteristics and biological functions. Biochimie. 2006;88:515–525. - PubMed
- Berglund J.A., Chua K., Abovich N., Reed R., Rosbash M., Chua K., Abovich N., Reed R., Rosbash M., Abovich N., Reed R., Rosbash M., Reed R., Rosbash M., Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources