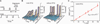Probing transcription factor dynamics at the single-molecule level in a living cell - PubMed (original) (raw)
Probing transcription factor dynamics at the single-molecule level in a living cell
Johan Elf et al. Science. 2007.
Abstract
Transcription factors regulate gene expression through their binding to DNA. In a living Escherichia coli cell, we directly observed specific binding of a lac repressor, labeled with a fluorescent protein, to a chromosomal lac operator. Using single-molecule detection techniques, we measured the kinetics of binding and dissociation of the repressor in response to metabolic signals. Furthermore, we characterized the nonspecific binding to DNA, one-dimensional (1D) diffusion along DNA segments, and 3D translocation among segments through cytoplasm at the single-molecule level. In searching for the operator, a lac repressor spends approximately 90% of time nonspecifically bound to and diffusing along DNA with a residence time of <5 milliseconds. The methods and findings can be generalized to other nucleic acid binding proteins.
Figures
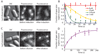
Fig. 1. Specific binding to lac operators
(A) Strains. The chromosomal lac region of the wild-type E. coli (BW25933) and various derivatives used in this report. (B) Bulk activity assay. The Miller assay (top) shows that the YFP fusion strains (JE12 and JE13) are active and respond to induction by IPTG (1 mM, 3 hours) by derepressing lacZ (yellow). The Western blot (bottom) for LacI shows that JE12 and JE13 express the full-length fusion protein (67kD) and that the expression in JE12, in the absence of IPTG, is strongly autorepressed as compared to the wild type and JE13. (C) Fluorescence (1-s exposure) and DIC images of JE12 grown in M9 glucose with amino acids. The YFP-labeled LacI binds persistently at one or two locations per cell depending on whether the operators have been replicated or not. The graph shows the fluorescence intensity along the red line. Scale bar, 1 µm. (D) DIC and fluorescence images (1-s exposure) of LacI-Venus expressed from plasmid in the lacI− and lacIOZ− strains, respectively. Plasmid expression is used to obtain similar expression levels in the two strains. No specific binding is observed in the absence of the lac operators.
Fig. 2. lac repressor kinetics in living cells
(A) JE12 bacteria before and 40 s after addition of IPTG to a final concentration of 1 mM. (B) Fraction of the lac operator regions that is TF-bound (±SEM, _n_~3) is plotted as a function of time after induction by various concentrations of IPTG. The data are globally fitted with a model in which IPTG binds independently to the two dimers in the operator region (SOM). (C) JE12 bacteria before and 1 min after dilution of IPTG from 100 to 2 µM with the addition of 1 mM ONPF (D) Fraction of the operator regions that is TF-bound (±SEM, _n_~3) as a function of time after rapid dilution of IPTG from 100 to 2 µM by addition of 1 mM ONPF. The data are fitted with an exponentially distributed binding time and yield a time constant of 59 s.
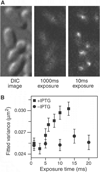
Fig. 3. Imaging nonspecifically bound LacI
(A) Two fluorescence images with different exposure times and the corresponding DIC images of IPTG-induced E. coli cells. At 1000 ms, individual LacI-Venus appear as diffuse fluorescence background. At 10 ms they are clearly visible as nearly diffraction-limited spots. (B) Fluorescence spot size as a function of exposure time. The size is represented as the average variance of a 2D Gaussian function fit to images of fluorescence spots (±SEM, _n_~100). The same total excitation energy is used for different exposure time. The spots are measured before (−IPTG, •) and after (+IPTG, ▪) induction. The size converges to the width of the point spread function (full width at half maximum = 370 nm) below 5 ms.
Fig. 4. Single molecule tracking with stroboscopic illumination
(A) Timing diagram for stroboscopic illumination. Each laser pulses (1 ms) is synchronized to the CCD frame time, which lasts time T. (B) Displacement histograms for different values of T. The absolute values of displacement along an arbitrary axis were calculated from 2D Gaussian fittings in two successive image frames. The displacement distribution of nonspecifically bound TFs broadens with time (left), whereas the distribution before induction (right) remains peaked at <100 nm. The contrast between them illustrates the change in TF mobility before and after induction. **(C)** Mean-square displacement for nonspecifically bound TFs at different time intervals. The red line shows a linear fit of the MSDs. Error bars are calculated as described in the SOM. The fitting agrees well with a normal diffusion in the imaging plane, <Δx2>=4Defft, with Deff=0.4 µm2s−1.
Similar articles
- The lac repressor displays facilitated diffusion in living cells.
Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. Hammar P, et al. Science. 2012 Jun 22;336(6088):1595-8. doi: 10.1126/science.1221648. Science. 2012. PMID: 22723426 - Biochemistry. Completing the view of transcriptional regulation.
von Hippel PH. von Hippel PH. Science. 2004 Jul 16;305(5682):350-2. doi: 10.1126/science.1101270. Science. 2004. PMID: 15256661 No abstract available. - DNA looping and lac repressor-CAP interaction.
Fried MG, Hudson JM. Fried MG, et al. Science. 1996 Dec 13;274(5294):1930-1; author reply 1931-2. doi: 10.1126/science.274.5294.1930. Science. 1996. PMID: 8984648 No abstract available. - The whole lactose repressor.
Matthews KS. Matthews KS. Science. 1996 Mar 1;271(5253):1245-6. doi: 10.1126/science.271.5253.1245. Science. 1996. PMID: 8638104 Review. No abstract available. - The lac repressor.
Lewis M. Lewis M. C R Biol. 2005 Jun;328(6):521-48. doi: 10.1016/j.crvi.2005.04.004. C R Biol. 2005. PMID: 15950160 Review.
Cited by
- Dissecting non-coding RNA mechanisms in cellulo by Single-molecule High-Resolution Localization and Counting.
Pitchiaya S, Krishnan V, Custer TC, Walter NG. Pitchiaya S, et al. Methods. 2013 Sep 15;63(2):188-99. doi: 10.1016/j.ymeth.2013.05.028. Epub 2013 Jun 29. Methods. 2013. PMID: 23820309 Free PMC article. - A benchmark for chromatin binding measurements in live cells.
Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. Mazza D, et al. Nucleic Acids Res. 2012 Aug;40(15):e119. doi: 10.1093/nar/gks701. Epub 2012 Jul 25. Nucleic Acids Res. 2012. PMID: 22844090 Free PMC article. - Real-time single-molecule imaging of transcriptional regulatory networks in living cells.
Hwang DW, Maekiniemi A, Singer RH, Sato H. Hwang DW, et al. Nat Rev Genet. 2024 Apr;25(4):272-285. doi: 10.1038/s41576-023-00684-9. Epub 2024 Jan 9. Nat Rev Genet. 2024. PMID: 38195868 Review. - Advances in Chromatin and Chromosome Research: Perspectives from Multiple Fields.
Agbleke AA, Amitai A, Buenrostro JD, Chakrabarti A, Chu L, Hansen AS, Koenig KM, Labade AS, Liu S, Nozaki T, Ovchinnikov S, Seeber A, Shaban HA, Spille JH, Stephens AD, Su JH, Wadduwage D. Agbleke AA, et al. Mol Cell. 2020 Sep 17;79(6):881-901. doi: 10.1016/j.molcel.2020.07.003. Epub 2020 Aug 7. Mol Cell. 2020. PMID: 32768408 Free PMC article. Review. - URDME: a modular framework for stochastic simulation of reaction-transport processes in complex geometries.
Drawert B, Engblom S, Hellander A. Drawert B, et al. BMC Syst Biol. 2012 Jun 22;6:76. doi: 10.1186/1752-0509-6-76. BMC Syst Biol. 2012. PMID: 22727185 Free PMC article.
References
- Matthews KS, Nichols JC. Prog Nucleic Acid Res Mol Biol. 1998;58:127. - PubMed
- Dunaway M, et al. J Biol Chem. 1980;255:10115. - PubMed
- Winter RB, von Hippel PH. Biochemistry. 1981;20:6948. - PubMed
- Lewis M, et al. Science. 1996;271:1247. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources