The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription - PubMed (original) (raw)
The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription
Deyarina Gonzalez et al. Mol Cell Biol. 2007 Aug.
Abstract
Transcription corepressors are general regulators controlling the expression of genes involved in multiple signaling pathways and developmental programs. Repression is mediated through mechanisms including the stabilization of a repressive chromatin structure over control regions and regulation of Mediator function inhibiting RNA polymerase II activity. Using whole-genome arrays we show that the Arabidopsis thaliana corepressor LEUNIG, a member of the GroTLE transcription corepressor family, regulates the expression of multiple targets in vivo. LEUNIG has a role in the regulation of genes involved in a number of different physiological processes including disease resistance, DNA damage response, and cell signaling. We demonstrate that repression of in vivo LEUNIG targets is achieved through histone deacetylase (HDAC)-dependent and -independent mechanisms. HDAC-dependent mechanisms involve direct interaction with HDA19, a class 1 HDAC, whereas an HDAC-independent repression activity involves interactions with the putative Arabidopsis Mediator components AtMED14/SWP and AtCDK8/HEN3. We suggest that changes in chromatin structure coupled with regulation of Mediator function are likely to be utilized by LEUNIG in the repression of gene transcription.
Figures
FIG. 1.
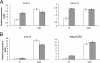
HDAC-dependent and -independent regulation of in vivo LUG targets. TSA treatment reveals HDAC-dependent (A) and HDAC-independent (B) LUG target genes. Shown are results of real-time PCR analysis of reverse-transcribed mRNA from Ler and lug-3 mutant seedlings 6 h post-TSA treatment. Relative quantification of gene expression data was carried out using CT values for each sample normalized to ACTIN2 expression levels. Normalized values for lug-3 expression levels are given as change (_n_-fold) relative to values for Ler seedlings.
FIG. 2.
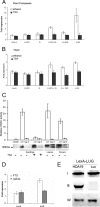
LUG interacts functionally and physically with HDACs to repress transcription. (A) LUG repression activity in plant protoplasts is abolished by TSA treatment. Arabidopsis leaf protoplasts were transfected with GAL4-LUG, GAL4-LUG derivatives, or GAL4-only effector plasmids plus reporter plasmid pJC1 and treated with 20 μM TSA (dark bars) or ethanol (white bars) for 6 h before fluorescence levels were determined relative to untransformed controls (blank). (B) LUG repression activity in yeast is abolished by TSA treatment. Yeast strain FT5 was transformed with the reporter vector pJK1621 (16) together with the indicated LexA-LUG derivatives (27) or LexA only. Individual transformants were treated with 20 μM TSA (dark bars) or ethanol (white bars) and grown overnight in liquid medium to early log phase (OD600 of <1) before β-galactosidase activity was determined. (C) LUG copurifies with an HDAC activity. Nuclear proteins isolated from Ler and lug-3 mutant leaves, flowers, and seedlings (with or without 6 h of TSA treatment) were incubated with immobilized anti-LUG antibody to immunopurify LUG-associated proteins, and the immunoprecipitate was either analyzed for HDAC activity relative to the negative bead-only control (top) or immunoblotted and probed with anti-LUG antibody (bottom). (D) LUG requires RPD3 to repress transcription. Yeast strains FT5 and FT5::_rpd3_Δ were transformed with the reporter vector pJK1621 (16) together with full-length LexA-LUG (27) or LexA only. Individual transformants were assayed as described for panel B. (E) LUG interacts with HDA19 in vitro. LexA-LUG was immunoprecipitated from yeast whole-cell extracts and incubated with [35S]methionine-labeled HDA19 or Luciferase (Luc). Input (I) and bound (B) 35S-labeled proteins were visualized using a phosphorimager. LUG was detected using an anti-LexA antibody (W).
FIG. 3.
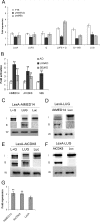
The Arabidopsis Mediator components AtMED14/SWP and AtCDK8/HEN3 are involved in LUG-mediated repression. (A) LUG requires yeast y_MED14_ and y_CDK8_ for repression function. Yeast strains FT5 (white bars), FT5::med14Δ (dark bars; viable partial deletion), and FT5::cdk8Δ (light gray bars) were transformed with the reporter vector pJK1621 (4× LexA operator) or the control vector pLG312S (0× LexA operator) together with the indicated LexA-LUG derivatives (27) or LexA only. Individual transformants were grown in liquid medium to an early log phase (OD600 of <1), and β-galactosidase activity was determined. Error bars show standard deviations. (B) LUG interacts with AtMED14 and AtCDK8 in vivo. Interactions were determined using yeast two-hybrid assays. FT5 was transformed with the reporter plasmid JK103 (4× LexA operator sequences-CYC1 minimal promoter-LacZ) plus LexA-AtMED14 or LexA-AtCDK8 together with either AD (AD only; white bars), LUG-AD (dark bars), or SEU-AD (light gray bars) (27). LexA-SEU was used as a positive control for LUG interaction. Error bars show standard deviations. Unpaired t tests comparing bait vector alone with bait plus prey vectors were used to test significance (**, P < 0.01). (C and E) LUG interacts with AtMED14 (C) and AtCDK8 (E) in vitro. LexA-MED14 or LexA-AtCDK8 was immunoprecipitated from whole-cell extracts and incubated with [35S]methionine-labeled LUG, LUFS+Q (L+Q), or luciferase (Luc) proteins. 35S-labeled proteins were visualized using a phosphorimager. LUG was detected using an anti-LexA antibody (W). (D and F) Like panels C and E but using LexA-LUG immunoprecipitated from yeast whole-cell extracts incubated with [35S]methionine-labeled AtMED14 (conserved N-terminal domain) (D) and AtCDK8 (F) or luciferase (Luc). I, input; B, bound. (G) AtMED14 and AtCDK8 display inherent repression activity. Repression activity of LexA-AtMED14 or LexA-AtCDK8 was determined by comparing LacZ activity in FT5 transformed with pJK1621 and activity in FT5 transformed with pLG312S, and significance was determined using unpaired t tests between pJK1621 and pLG312S values (**, P < 0.01). Error bars show standard deviations.
Similar articles
- Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2.
Conlan RS, Tzamarias D. Conlan RS, et al. J Mol Biol. 2001 Jun 22;309(5):1007-15. doi: 10.1006/jmbi.2001.4742. J Mol Biol. 2001. PMID: 11399075 - Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms.
Sun H, Taneja R. Sun H, et al. Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):4058-63. doi: 10.1073/pnas.070526297. Proc Natl Acad Sci U S A. 2000. PMID: 10737769 Free PMC article. - A mechanism of COOH-terminal binding protein-mediated repression.
Meloni AR, Lai CH, Yao TP, Nevins JR. Meloni AR, et al. Mol Cancer Res. 2005 Oct;3(10):575-83. doi: 10.1158/1541-7786.MCR-05-0088. Mol Cancer Res. 2005. PMID: 16254191 - Corepressor requirement and thyroid hormone receptor function during Xenopus development.
Sachs LM. Sachs LM. Vitam Horm. 2004;68:209-30. doi: 10.1016/S0083-6729(04)68007-1. Vitam Horm. 2004. PMID: 15193456 Review. - Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor.
Lutz M, Baniahmad A, Renkawitz R. Lutz M, et al. Biochem Soc Trans. 2000;28(4):386-9. Biochem Soc Trans. 2000. PMID: 10961925 Review.
Cited by
- Analysis of differential expression of Mediator subunit genes in Arabidopsis.
Pasrija R, Thakur JK. Pasrija R, et al. Plant Signal Behav. 2012 Dec;7(12):1676-86. doi: 10.4161/psb.22438. Epub 2012 Oct 16. Plant Signal Behav. 2012. PMID: 23072992 Free PMC article. - The Transcriptional Coregulator LEUNIG_HOMOLOG Inhibits Light-Dependent Seed Germination in Arabidopsis.
Lee N, Park J, Kim K, Choi G. Lee N, et al. Plant Cell. 2015 Aug;27(8):2301-13. doi: 10.1105/tpc.15.00444. Epub 2015 Aug 14. Plant Cell. 2015. PMID: 26276832 Free PMC article. - LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development.
Sitaraman J, Bui M, Liu Z. Sitaraman J, et al. Plant Physiol. 2008 Jun;147(2):672-81. doi: 10.1104/pp.108.115923. Epub 2008 Apr 4. Plant Physiol. 2008. PMID: 18390806 Free PMC article. - The role of auxin in style development and apical-basal patterning of the Arabidopsis thaliana gynoecium.
Ståldal V, Sundberg E. Ståldal V, et al. Plant Signal Behav. 2009 Feb;4(2):83-5. doi: 10.4161/psb.4.2.7538. Plant Signal Behav. 2009. PMID: 19649177 Free PMC article. Review. - The TOPLESS interactome: a framework for gene repression in Arabidopsis.
Causier B, Ashworth M, Guo W, Davies B. Causier B, et al. Plant Physiol. 2012 Jan;158(1):423-38. doi: 10.1104/pp.111.186999. Epub 2011 Nov 7. Plant Physiol. 2012. PMID: 22065421 Free PMC article.
References
- Asamizu, E., Y. Nakamura, S. Sato, and S. Tabata. 2000. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7:175-180. - PubMed
- Boube, M., L. Joulia, D. L. Cribbs, and H. M. Bourbon. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. - PubMed
- Bourbon, H. M., A. Aguilera, A. Z. Ansari, F. J. Asturias, A. J. Berk, S. Bjorklund, T. K. Blackwell, T. Borggrefe, M. Carey, M. Carlson, J. W. Conaway, R. C. Conaway, S. W. Emmons, J. D. Fondell, L. P. Freedman, T. Fukasawa, C. M. Gustafsson, M. Han, X. He, P. K. Herman, A. G. Hinnebusch, S. Holmberg, F. C. Holstege, J. A. Jaehning, Y. J. Kim, L. Kuras, A. Leutz, J. T. Lis, M. Meisterernest, A. M. Naar, K. Nasmyth, J. D. Parvin, M. Ptashne, D. Reinberg, H. Ronne, I. Sadowski, H. Sakurai, M. Sipiczki, P. W. Sternberg, D. J. Stillman, R. Strich, K. Struhl, J. Q. Svejstrup, S. Tuck, F. Winston, R. G. Roeder, and R. D. Kornberg. 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14:553-557. - PubMed
- Cnops, G., S. Jover-Gil, J. L. Peters, P. Neyt, S. De Block, P. Robles, M. R. Ponce, T. Gerats, J. L. Micol, and M. Van Lijsebettens. 2004. The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. J. Exp. Bot. 55:1529-1539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases