The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling - PubMed (original) (raw)
The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling
Yannick Jacob et al. Plant Physiol. 2007 Jul.
Abstract
Nuclear pore complexes (NPCs) mediate the transport of RNA and other cargo between the nucleus and the cytoplasm. In vertebrates, the NPC protein TRANSLOCATED PROMOTER REGION (TPR) is associated with the inner filaments of the nuclear basket and is thought to serve as a scaffold for the assembly of transport machinery. In a screen for mutants that suppress the expression of the floral inhibitor FLOWERING LOCUS C, we identified lesions in the Arabidopsis (Arabidopsis thaliana) homolog of TPR (AtTPR). attpr mutants exhibit early-flowering and other pleiotropic phenotypes. A possible explanation for these developmental defects is that attpr mutants exhibit an approximately 8-fold increase in nuclear polyA RNA. Thus AtTPR is required for the efficient export of RNA from the nucleus. Microarray analysis shows that, in wild type, transcript abundance in the nuclear and total RNA pools are highly correlated; whereas, in attpr mutants, a significantly larger fraction of transcripts is enriched in either the nuclear or total pool. Thus AtTPR is required for homeostasis between nuclear and cytoplasmic RNA. We also show that the effects of AtTPR on small RNA abundance and auxin signaling are similar to that of two other NPC-associated proteins, HASTY (HST) and SUPPRESSOR OF AUXIN RESISTANCE3 (SAR3). This suggests that AtTPR, HST, and SAR3 may play related roles in the function of the nuclear pore.
Figures
Figure 1.
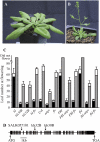
attpr mutations and their effect on flowering time. _ld_-1 (A) and lds30B (B) grown in long days for 4 weeks. C, The effect of attpr mutations on flowering time in the indicated backgrounds. Black and gray bars represent the number of primary rosette leaves formed prior to flowering in long and short days. The white portions of the bars represent the number of cauline leaves. Error bars indicate one
sd
. D, A schematic representation of AtTPR; the thin line indicates the chromosome, thick lines indicate exons. The positions of T-DNA insertions in attpr mutants are indicated above the gene. [See online article for color version of this figure.]
Figure 2.
Effect of attpr on development and gene expression. A, Rosette leaves formed prior to flowering in long days by Col (top) and attpr (bottom). B to G, Scanning electron micrographs of Col (B, D, and F) and attpr (C, E, and G) inflorescences (B and C) and sepals (D–G). White bars represent 1 mm (B and C), 100 _μ_m (D and E), and 10 _μ_m (F and G). H, RT-PCR analysis of gene expression in the indicated genotypes; numbers indicate fold changes relative to Col. RNA was isolated from 2-week-old seedlings grown under long days. UBQ10 was used as a constitutively expressed control. [See online article for color version of this figure.]
Figure 3.
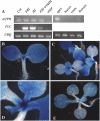
AtTPR expression. A, RT-PCR analysis of gene expression in the indicated genotypes and tissues. UBQ10 was used as a constitutively expressed control. B to E, Histochemical expression of GUS expression in AtTPR∷GUS transgenic plants.
Figure 4.
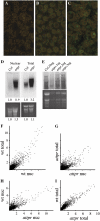
RNA analysis in attpr mutants. A to C, Whole-mount in situ localization of polyA RNA. Confocal images of leaves of Col (A and B) and attpr (C) hybridized without (A) or with (B and C) a poly(dT) fluorescein-tagged oligonucleotide. Leaves were harvested from 3-week-old plants grown under short days (A–C). D and E, RNA-blot analysis of polyA RNA in nuclear and total RNA from Col and attpr. Blots were probed with a radiolabeled poly(dT) oligonucleotide. D, Numbers indicate fold changes relative to Col. E, Size distribution of polyA RNA in Col and attpr total RNA. Photographs of the ethidium-stained gels used to produce the RNA blots are shown in the bottom sections (D and E) and were used as a control for loading. F to I, Microarray analysis in Col and the attpr mutant. Scatter plots of normalized signal intensities are shown for the indicated samples.
Figure 5.
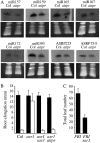
Effects of attpr on small RNA abundance and auxin signaling. A, RNA-blot analysis of small RNAs. Total RNA was isolated from the above-ground portions of long-day-grown flowering plants. B, Root elongation after 4 d on auxin-containing media for the indicated genotypes. C, Effect of sar3 on flowering time in a _FRI_-containing background. B and C, Error bars indicate one
sd
.
Similar articles
- NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development.
Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. Xu XM, et al. Plant Cell. 2007 May;19(5):1537-48. doi: 10.1105/tpc.106.049239. Epub 2007 May 18. Plant Cell. 2007. PMID: 17513499 Free PMC article. - Peering through the pore: The role of AtTPR in nuclear transport and development.
Jacob Y, Michaels SD. Jacob Y, et al. Plant Signal Behav. 2008 Jan;3(1):62-4. doi: 10.4161/psb.3.1.4903. Plant Signal Behav. 2008. PMID: 19704774 Free PMC article. - The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development.
Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. Parry G, et al. Plant Cell. 2006 Jul;18(7):1590-603. doi: 10.1105/tpc.106.041566. Epub 2006 Jun 2. Plant Cell. 2006. PMID: 16751346 Free PMC article. - Nuclear RNA export and its importance in abiotic stress responses of plants.
Chinnusamy V, Gong Z, Zhu JK. Chinnusamy V, et al. Curr Top Microbiol Immunol. 2008;326:235-55. doi: 10.1007/978-3-540-76776-3_13. Curr Top Microbiol Immunol. 2008. PMID: 18630756 Review. - Assessing the function of the plant nuclear pore complex and the search for specificity.
Parry G. Parry G. J Exp Bot. 2013 Feb;64(4):833-45. doi: 10.1093/jxb/ers289. Epub 2012 Oct 17. J Exp Bot. 2013. PMID: 23077202 Review.
Cited by
- Technologies for systems-level analysis of specific cell types in plants.
Wang D, Mills ES, Deal RB. Wang D, et al. Plant Sci. 2012 Dec;197:21-29. doi: 10.1016/j.plantsci.2012.08.012. Epub 2012 Aug 30. Plant Sci. 2012. PMID: 23116668 Free PMC article. Review. - Nucleo-cytoplasmic transport of proteins and RNA in plants.
Merkle T. Merkle T. Plant Cell Rep. 2011 Feb;30(2):153-76. doi: 10.1007/s00299-010-0928-3. Epub 2010 Oct 20. Plant Cell Rep. 2011. PMID: 20960203 Free PMC article. Review. - The nucleoporin NUP160 and NUP96 regulate nucleocytoplasmic export of mRNAs and participate in ethylene signaling and response in Arabidopsis.
Nie Y, Li Y, Liu M, Ma B, Sui X, Chen J, Yu Y, Dong CH. Nie Y, et al. Plant Cell Rep. 2023 Mar;42(3):549-559. doi: 10.1007/s00299-022-02976-6. Epub 2023 Jan 4. Plant Cell Rep. 2023. PMID: 36598573 - Endosperm-specific transcriptome analysis by applying the INTACT system.
Del Toro-De León G, Köhler C. Del Toro-De León G, et al. Plant Reprod. 2019 Mar;32(1):55-61. doi: 10.1007/s00497-018-00356-3. Epub 2018 Dec 26. Plant Reprod. 2019. PMID: 30588542 - The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana.
Deal RB, Henikoff S. Deal RB, et al. Nat Protoc. 2011 Jan;6(1):56-68. doi: 10.1038/nprot.2010.175. Epub 2010 Dec 16. Nat Protoc. 2011. PMID: 21212783 Free PMC article.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
- Bangs PL, Sparks CA, Odgren PR, Fey EG (1996) Product of the oncogene-activating gene Tpr is a phosphorylated protein of the nuclear pore complex. J Cell Biochem 61 48–60 - PubMed
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM (2004) Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J 40 112–119 - PubMed
- Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of EXPORTIN 5/MSN5, regulates phase change and morphogenesis. Development 130 1493–1504 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials