Formation of native prions from minimal components in vitro - PubMed (original) (raw)
Comparative Study
. 2007 Jun 5;104(23):9741-6.
doi: 10.1073/pnas.0702662104. Epub 2007 May 29.
Affiliations
- PMID: 17535913
- PMCID: PMC1887554
- DOI: 10.1073/pnas.0702662104
Comparative Study
Formation of native prions from minimal components in vitro
Nathan R Deleault et al. Proc Natl Acad Sci U S A. 2007.
Erratum in
- Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12636
Abstract
The conformational change of a host protein, PrP(C), into a disease-associated isoform, PrP(Sc), appears to play a critical role in the pathogenesis of prion diseases such as Creutzfeldt-Jakob disease and scrapie. However, the fundamental mechanism by which infectious prions are produced in neurons remains unknown. To investigate the mechanism of prion formation biochemically, we conducted a series of experiments using the protein misfolding cyclic amplification (PMCA) technique with a preparation containing only native PrP(C) and copurified lipid molecules. These experiments showed that successful PMCA propagation of PrP(Sc) molecules in a purified system requires accessory polyanion molecules. In addition, we found that PrP(Sc) molecules could be formed de novo from these defined components in the absence of preexisting prions. Inoculation of samples containing either prion-seeded or spontaneously generated PrP(Sc) molecules into hamsters caused scrapie, which was transmissible on second passage. These results show that prions able to infect wild-type hamsters can be formed from a minimal set of components including native PrP(C) molecules, copurified lipid molecules, and a synthetic polyanion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
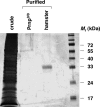
Silver stain analysis of purified PrPC substrate. Twelve percent SDS/PAGE showing (from left to right): crude, detergent-solubilized brain supernatant; preparation mock-purified from Prnp0/0 mouse brains; preparation purified from normal hamster brains; and molecular weight markers.
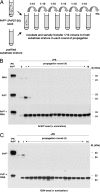
Fig. 2.
Seeded PrPSc propagation by using purified substrates. (A) Schematic diagram of serial dilution and propagation paradigm used in subsequent experiments, adapted from Castilla et al. (22). (B and C) Western blots showing samples subjected to 16 rounds of PMCA, serial dilution, and propagation as depicted in A. (B) Samples originally seeded with Sc237 PrP27-30 were propagated in substrate containing either poly(A) RNA alone (top gel), purified PrPC alone (middle gel), or PrPC plus poly(A) RNA (bottom gel). (C) Samples originally seeded with 139H PrP27-30 were propagated in substrate containing either purified PrPC alone (top gel), or poly(A) PrPC plus poly(A) RNA (bottom gel). In all gels, a sample containing PrPC not subjected to proteinase K digestion is shown in the first lane as a reference for comparison of electrophoretic mobility (PrPC-PK). All other samples were subjected to limited proteolysis with 50 μg/ml proteinase K (+PK).
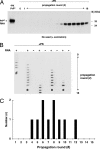
Fig. 3.
Formation of PrPSc molecules de novo during serial PMCA propagation of unseeded purified substrates. (A) Western blot showing unseeded samples containing PrPC plus poly(A) RNA subjected to 16 rounds of PMCA, serial dilution, and propagation. A sample containing PrPC not subjected to proteinase K digestion is shown in the first lane as a reference for comparison of electrophoretic mobility (PrPC-PK). All other samples were subjected to limited proteolysis with 50 μg/ml proteinase K (+PK). (B) Representative slot blot showing the formation of protease-resistant PrP molecules de novo in multiple propagation experiments. Unseeded samples were propagated for 15 rounds in either the presence or absence of poly(A) RNA, as indicated by plus (+) and minus (−) symbols, respectively. In experiments designated by asterisks, propagation was not carried beyond the 12th round because a preliminary assay performed at that stage already detected PrPSc in several preceding rounds. The experiment designated by the # symbol was performed entirely in a prion-free laboratory. (C) Histogram showing the temporal distribution of the first detectable PrPSc signals during serial propagation of unseeded PrPC plus poly(A) RNA substrate mixtures in 10 separate propagation experiments, 7 of which were carried out simultaneously in the same sonicator.
Fig. 4.
Representative histological fields of the CA2 hippocampus region in control animals and animals inoculated with _in vitro_-generated PrPSc molecules. Rows from top to bottom: (top row) uninoculated, normal 190 day old hamster; (second row) terminally ill hamster inoculated with Sc237-seeded, serially propagated PrPSc molecules; (third row) terminally ill hamster inoculated with 139H-seeded, serially propagated PrPSc molecules; (bottom row) terminally ill hamster inoculated with spontaneously generated, serially propagated PrPSc molecules. Hematoxylin and eosin (H&E) staining, as well as glial fibrillary acidic protein (GFAP) and PrP immunohistochemical staining results are shown for each group. (Scale bar, 100 μm.)
Comment in
- A simplified recipe for prions.
Lee KS, Caughey B. Lee KS, et al. Proc Natl Acad Sci U S A. 2007 Jun 5;104(23):9551-2. doi: 10.1073/pnas.0703910104. Epub 2007 May 29. Proc Natl Acad Sci U S A. 2007. PMID: 17535888 Free PMC article. No abstract available.
Similar articles
- PrP(Sc) of scrapie 263K propagates efficiently in spleen and muscle tissues with protein misfolding cyclic amplification.
Shi S, Dong CF, Wang GR, Wang X, An R, Chen JM, Shan B, Zhang BY, Xu K, Shi Q, Tian C, Gao C, Han J, Dong XP. Shi S, et al. Virus Res. 2009 Apr;141(1):26-33. doi: 10.1016/j.virusres.2008.12.010. Epub 2009 Jan 20. Virus Res. 2009. PMID: 19162101 - Generation of genuine prion infectivity by serial PMCA.
Weber P, Giese A, Piening N, Mitteregger G, Thomzig A, Beekes M, Kretzschmar HA. Weber P, et al. Vet Microbiol. 2007 Aug 31;123(4):346-57. doi: 10.1016/j.vetmic.2007.04.004. Epub 2007 Apr 7. Vet Microbiol. 2007. PMID: 17493773 - Prion encephalopathies of animals and humans.
Prusiner SB. Prusiner SB. Dev Biol Stand. 1993;80:31-44. Dev Biol Stand. 1993. PMID: 8270114 Review. - Autocatalytic self-propagation of misfolded prion protein.
Bieschke J, Weber P, Sarafoff N, Beekes M, Giese A, Kretzschmar H. Bieschke J, et al. Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12207-11. doi: 10.1073/pnas.0404650101. Epub 2004 Aug 5. Proc Natl Acad Sci U S A. 2004. PMID: 15297610 Free PMC article. - Prion protein conversion in vitro.
Supattapone S. Supattapone S. J Mol Med (Berl). 2004 Jun;82(6):348-56. doi: 10.1007/s00109-004-0534-3. Epub 2004 Mar 10. J Mol Med (Berl). 2004. PMID: 15014886 Review.
Cited by
- Rapid and sensitive determination of residual prion infectivity from prion-decontaminated surfaces.
Simmons SM, Payne VL, Hrdlicka JG, Taylor J, Larsen PA, Wolf TM, Schwabenlander MD, Yuan Q, Bartz JC. Simmons SM, et al. mSphere. 2024 Sep 25;9(9):e0050424. doi: 10.1128/msphere.00504-24. Epub 2024 Aug 27. mSphere. 2024. PMID: 39189773 Free PMC article. - Neuropathologically directed profiling of PRNP somatic and germline variants in sporadic human prion disease.
McDonough GA, Cheng Y, Morillo KS, Doan RN, Zhou Z, Kenny CJ, Foutz A, Kim C, Cohen ML, Appleby BS, Walsh CA, Safar JG, Huang AY, Miller MB. McDonough GA, et al. Acta Neuropathol. 2024 Jul 24;148(1):10. doi: 10.1007/s00401-024-02774-2. Acta Neuropathol. 2024. PMID: 39048735 - Neuropathologically-directed profiling of PRNP somatic and germline variants in sporadic human prion disease.
McDonough GA, Cheng Y, Morillo K, Doan RN, Kenny CJ, Foutz A, Kim C, Cohen ML, Appleby BS, Walsh CA, Safar JG, Huang AY, Miller MB. McDonough GA, et al. bioRxiv [Preprint]. 2024 Jun 29:2024.06.25.600668. doi: 10.1101/2024.06.25.600668. bioRxiv. 2024. PMID: 38979287 Free PMC article. Updated. Preprint. - Roles of Nucleic Acids in Protein Folding, Aggregation, and Disease.
Litberg TJ, Horowitz S. Litberg TJ, et al. ACS Chem Biol. 2024 Apr 19;19(4):809-823. doi: 10.1021/acschembio.3c00695. Epub 2024 Mar 13. ACS Chem Biol. 2024. PMID: 38477936 Review. - A Protein Misfolding Shaking Amplification-based method for the spontaneous generation of hundreds of bona fide prions.
Eraña H, Sampedro-Torres-Quevedo C, Charco JM, Díaz-Domínguez CM, Peccati F, San-Juan-Ansoleaga M, Vidal E, Gonçalves-Anjo N, Pérez-Castro MA, González-Miranda E, Piñeiro P, Fernández-Veiga L, Galarza-Ahumada J, Fernández-Muñoz E, Perez de Nanclares G, Telling G, Geijo M, Jiménez-Osés G, Castilla J. Eraña H, et al. Nat Commun. 2024 Mar 8;15(1):2112. doi: 10.1038/s41467-024-46360-2. Nat Commun. 2024. PMID: 38459071 Free PMC article.
References
- Prusiner SB. Science. 1982;216:136–144. - PubMed
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Science. 2004;305:673–676. - PubMed
- Sparrer HE, Santoso A, Szoka FC, Jr, Weissman JS. Science. 2000;289:595–599. - PubMed
- Telling GC, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen FE, DeArmond SJ, Prusiner SB. Cell. 1995;83:79–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials