A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites - PubMed (original) (raw)
A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites
Dorothée Molle et al. Retrovirology. 2007.
Abstract
HIV-1 transcription is tightly regulated: silent in long-term latency and highly active in acutely-infected cells. Transcription is activated by the viral protein Tat, which recruits the elongation factor P-TEFb by binding the TAR sequence present in nascent HIV-1 RNAs. In this study, we analyzed the dynamic of the TAR:Tat:P-TEFb complex in living cells, by performing FRAP experiments at HIV-1 transcription sites. Our results indicate that a large fraction of Tat present at these sites is recruited by Cyclin T1. We found that in the presence of Tat, Cdk9 remained bound to nascent HIV-1 RNAs for 71s. In contrast, when transcription was activated by PMA/ionomycin, in the absence of Tat, Cdk9 turned-over rapidly and resided on the HIV-1 promoter for only 11s. Thus, the mechanism of trans-activation determines the residency time of P-TEFb at the HIV-1 gene, possibly explaining why Tat is such a potent transcriptional activator. In addition, we observed that Tat occupied HIV-1 transcription sites for 55s, suggesting that the TAR:Tat:P-TEFb complex dissociates from the polymerase following transcription initiation, and undergoes subsequent cycles of association/dissociation.
Figures
Figure 1
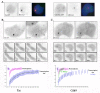
Dynamic of Tat and Cdk9 at HIV-1 transcription sites. A-Accumulation of Tat, but not the C22G mutant, at HIV-1 transcription sites. U2OS_HIV-1 cells were transfected with Tat-GFP or Tat(C22G)-GFP, and then induced 7h with PMA/ionomycin. Cells were then fixed and hybridized in situ with a Cy3-labelled oligo probe against the MS2 repeat. The HIV-1 transcription site corresponds to the focal accumulation labelled by the MS2 probe. Blue: dapi. Each field is 22 × 22 μm. B-Dynamic of Tat at HIV-1 transcription sites. U2OS_HIV-1 cells were transfected with vectors expressing Tat-CFP and MS2-YFP. Tat-CFP was then bleached, and recovery was analyzed by tracking transcription sites in 3D with a wide-field microscope. Upper panel: colocalization of Tat-CFP and MS2-YFP in living cells (30 × 25μm). Middle panels: image sequence from a FRAP experiment (time in second; each field is 30 × 25 μm). Graph: recovery curves in the nucleoplasm of transfected U2OS cells (pink), or at the HIV-1 transcription site (blue). The best fit is shown in green. C-Dynamics of Cdk9 at HIV-1 transcription sites. U2OS_HIV-1 cells were transfected with vectors expressing Cdk9-GFP and MS2-mCherry. Cdk9-GFP was then bleached, and recovery was analyzed by tracking transcription sites in 3D with a wide-field microscope. Upper panel: colocalization of Cdk9-GFP and MS2-mCherry in living cells (30 × 25 μm). Middle panels: image sequence from a FRAP experiment (time in second; each field is 30 × 25 μm). Graph: recovery curves in the nucleoplasm of transfected U2OS cells (pink), or at the HIV-1 transcription site. Blue: cells were transfected with Tat; Green: Tat was absent but cells were induced by PMA/ionomycin.
Figure 2
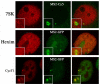
Hexim and 7SK are not recruited at the HIV-1 transcription site. U2OS_HIV-1 cells were transfected with vectors expressing Tat alone (upper panel), or Tat and MS2-GFP (middle and lower panels). 24h later, cells were fixed and processed. Upper panel: cells were hybridized in situ with fluorescent oligonucleotide probes detecting 7SK (red) or the MS2 repeat (green). Middle and lower panels, cells were labeled with antibodies against Hexim1 (red, middle panel), or cyclin T1 (red, lower panel).
Similar articles
- HIV-1 tat transcriptional activity is regulated by acetylation.
Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C. Kiernan RE, et al. EMBO J. 1999 Nov 1;18(21):6106-18. doi: 10.1093/emboj/18.21.6106. EMBO J. 1999. PMID: 10545121 Free PMC article. - Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages.
Zhou Q, Chen D, Pierstorff E, Luo K. Zhou Q, et al. EMBO J. 1998 Jul 1;17(13):3681-91. doi: 10.1093/emboj/17.13.3681. EMBO J. 1998. PMID: 9649438 Free PMC article. - Tackling Tat.
Karn J. Karn J. J Mol Biol. 1999 Oct 22;293(2):235-54. doi: 10.1006/jmbi.1999.3060. J Mol Biol. 1999. PMID: 10550206 Review. - Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression.
Romano G, Kasten M, De Falco G, Micheli P, Khalili K, Giordano A. Romano G, et al. J Cell Biochem. 1999 Dec 1;75(3):357-68. J Cell Biochem. 1999. PMID: 10536359 Review.
Cited by
- Functional roles of HIV-1 Tat protein in the nucleus.
Musinova YR, Sheval EV, Dib C, Germini D, Vassetzky YS. Musinova YR, et al. Cell Mol Life Sci. 2016 Feb;73(3):589-601. doi: 10.1007/s00018-015-2077-x. Epub 2015 Oct 27. Cell Mol Life Sci. 2016. PMID: 26507246 Free PMC article. Review. - Live-Cell Imaging of Early Steps of Single HIV-1 Infection.
Francis AC, Melikyan GB. Francis AC, et al. Viruses. 2018 May 19;10(5):275. doi: 10.3390/v10050275. Viruses. 2018. PMID: 29783762 Free PMC article. Review. - Stochastic pausing at latent HIV-1 promoters generates transcriptional bursting.
Tantale K, Garcia-Oliver E, Robert MC, L'Hostis A, Yang Y, Tsanov N, Topno R, Gostan T, Kozulic-Pirher A, Basu-Shrivastava M, Mukherjee K, Slaninova V, Andrau JC, Mueller F, Basyuk E, Radulescu O, Bertrand E. Tantale K, et al. Nat Commun. 2021 Jul 23;12(1):4503. doi: 10.1038/s41467-021-24462-5. Nat Commun. 2021. PMID: 34301927 Free PMC article. - Visual detection of Flavivirus RNA in living cells.
Miorin L, Maiuri P, Marcello A. Miorin L, et al. Methods. 2016 Apr 1;98:82-90. doi: 10.1016/j.ymeth.2015.11.002. Epub 2015 Nov 2. Methods. 2016. PMID: 26542763 Free PMC article. Review. - Spatial Organization and Dynamics of Transcription Elongation and Pre-mRNA Processing in Live Cells.
Sánchez-Álvarez M, Sánchez-Hernández N, Suñé C. Sánchez-Álvarez M, et al. Genet Res Int. 2011;2011:626081. doi: 10.4061/2011/626081. Epub 2011 Nov 24. Genet Res Int. 2011. PMID: 22567362 Free PMC article.
References
- Marcello A, Zoppe M, Giacca M. Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life. 2001;51:175–181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous