A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein - PubMed (original) (raw)
A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein
Yasushi Muraki et al. J Virol. 2007 Aug.
Abstract
Reverse genetics has been documented for influenza A, B, and Thogoto viruses belonging to the family Orthomyxoviridae. We report here the reverse genetics of influenza C virus, another member of this family. The seven viral RNA (vRNA) segments of C/Ann Arbor/1/50 were expressed in 293T cells from cloned cDNAs, together with nine influenza C virus proteins. At 48 h posttransfection, the infectious titer of the culture supernatant was determined to be 2.51 x 10(3) 50% egg infectious doses/ml, which is lower than the number of influenza C virus-like particles (VLPs) (10(6)/ml) generated using the same system. By generating influenza C VLPs containing a given vRNA segment, we showed that each of the vRNA segments was similarly synthesized in the plasmid-transfected cells but that some segments were less efficiently incorporated into the VLPs. This finding leads us to speculate that the differences in incorporation efficiency into VLPs between segments might be a reason for the inefficient production of infectious viruses. Second, we generated a mutant recombinant virus, rMG96A, which possesses an Ala-->Thr mutation at residue 24 of the M1 protein, a substitution demonstrated to be involved in the morphology (filamentous or spherical) of the influenza C VLPs. As expected, rMG96A exhibited a spherical morphology, whereas recombinant wild-type of C/Ann Arbor/1/50, rWT, exhibited a mainly filamentous morphology. Membrane flotation analysis of the cells infected with rWT or rMG96A revealed a difference in the ratio of membrane-associated M1 proteins, suggesting that the affinity of M1 protein to the cell membrane is a determinant for virion morphology.
Figures
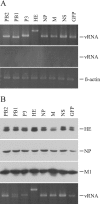
FIG. 1.
Generation of influenza C VLP containing a given vRNA segment. 293T cells were transfected with one of the indicated PolI plasmids [pPolI/PB2, pPolI/PB1, pPolI/P3, pPolI/HE, pPolI/NP, pPolI/M, pPolI/NS, or pPolI/NP-AA.GFP(−)], together with the nine virus protein-expressing plasmids. A representative result from three independent experiments is shown. (A) At 48 h p.t., RNA was extracted from the cytoplasm of the transfected cells, treated with DNase, reverse transcribed, and then PCR amplified with a set of primer specific for each gene segment. The PCR products were electrophoresed in an agarose gel (upper panel). To rule out the possibility that the RNA preparation was contaminated with residual plasmids, preparations without reverse transcription were used for PCR (middle panel). As an internal control, aliquots of extracted RNA were each applied to reverse transcribed-PCR for chicken β-actin mRNA (lower panel). The number of PCR cycles for chicken β-actin mRNA was adjusted to 18, at which number the amount of the PCR product increased in a log phase. To detect vRNAs, the amount of template from each construct was adjusted according to the ratio of β-actin PCR product, and then PCR (18 cycles) was performed. The sizes of the obtained PCR products were 505 bp for the PB2 gene, 482 bp for the PB1 gene, 528 bp for the P3 gene, 711 bp for the HE gene, 509 bp for the NP gene, 508 bp for the M gene, 502 bp for the NS gene, and 504 bp for the GFP gene. (B) At 48 h p.t., influenza C VLPs were collected from the culture medium and analyzed by Western blotting with antiserum against AA/50 virions (upper three panels). The RNA was extracted from the aliquots of the VLP preparation, treated with DNase, reverse transcribed, and then PCR amplified with a set of primer specific for each gene segment (lowest panel).
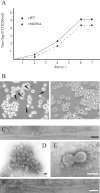
FIG. 2.
Infection experiments with recombinant viruses. (A) Growth kinetics of recombinant viruses. HMV-II cells were infected with rWT or rMG96A at a multiplicity of infection of 0.005 TCID50 and then incubated in the presence of trypsin (20 μg/ml). At 4, 6, and 7 days p.i., the culture media were used for the determination of infectious titers (TCID50/ml, see the text). A representative result of three independent experiments is shown. (B) CLS on the infected cells. HMV-II cells were infected with rWT (left panel) or rMG96A (right panel) as described above, and the cells were observed under a light microscope (magnification, ×350) at 4 days p.i. The CLS protruding from the cells are indicated as triangles. (C to F) Electron micrographs of recombinant viruses. The rWT grown in HMV-II cells (C) or the amniotic cavity of chicken eggs (F) and the rMG96A grown in eggs (D) or HMV-II cells (E) were concentrated, negatively stained, and examined under an electron microscope. Bar, 100 nm.
FIG. 3.
Membrane flotation analyses of infected or transfected cells. (A) HMV-II cells were infected with rWT or rMG96A at a multiplicity of infection of 5 TCID50, and the postnuclear supernatants were subjected to flotation analysis at 20 h p.i. as described in Materials and Methods. Virus proteins in each fraction were analyzed by Western blotting with antiserum against the AA/50 virion. Membrane-bound material floats up to the interface between 10 and 65% sucrose (fractions 3 and 4). A representative result of three independent experiments is shown. (B and C) HMV-II cells infected with rWT or rMG96A were pulse-labeled with [35S]methionine for 15 min at 20 h p.i. (B) and chased for 2.5 h (C). The cells were subjected to flotation analysis. Virus proteins in each fraction were immunoprecipitated with antiserum against the AA/50 virion and analyzed by SDS-PAGE. A representative result of two independent experiments is shown. (D) 293T cells transfected with pCAGGS.MCS/M1-AA or pCAGGS.MCS/M1-TAY were subjected to flotation analysis and analyzed by Western blotting. The latter plasmid expresses the M1 of C/Taylor/1233/47, which has a Thr at residue 24.
FIG. 4.
Raft association of virus proteins. HMV-II cells infected with rWT (A) or rMG96A (B) were pulse-labeled and chased for 4 h. The cells were treated with an NTE buffer containing 0.45% Triton X-100 on ice, and the lysates were subjected to flotation analysis. Virus proteins in each fraction were immunoprecipitated with antiserum against the AA/50 virion and analyzed by SDS-PAGE. (C) HMV-II cells infected with A/Aichi/2/68 were pulse-labeled for 15 min at 10 h p.i. and chased for 1.5 h. After fractionation, the HA protein in each fraction was immunoprecipitated with antiserum against the A/Aichi/2/68 virion and analyzed.
Similar articles
- Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins.
Latham T, Galarza JM. Latham T, et al. J Virol. 2001 Jul;75(13):6154-65. doi: 10.1128/JVI.75.13.6154-6165.2001. J Virol. 2001. PMID: 11390617 Free PMC article. - The molecular virology and reverse genetics of influenza C virus.
Muraki Y, Hongo S. Muraki Y, et al. Jpn J Infect Dis. 2010 May;63(3):157-65. Jpn J Infect Dis. 2010. PMID: 20495266 Review. - Palmitoylation of CM2 is dispensable to influenza C virus replication.
Muraki Y, Okuwa T, Furukawa T, Matsuzaki Y, Sugawara K, Himeda T, Hongo S, Ohara Y. Muraki Y, et al. Virus Res. 2011 Apr;157(1):99-105. doi: 10.1016/j.virusres.2011.02.013. Epub 2011 Feb 23. Virus Res. 2011. PMID: 21352864 - [Type C influenza].
Hongo S. Hongo S. Nihon Rinsho. 2006 Oct;64(10):1942-9. Nihon Rinsho. 2006. PMID: 17037372 Review. Japanese.
Cited by
- Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses-A Comparative Review on Influenza C and D.
Sreenivasan CC, Sheng Z, Wang D, Li F. Sreenivasan CC, et al. Pathogens. 2021 Dec 5;10(12):1583. doi: 10.3390/pathogens10121583. Pathogens. 2021. PMID: 34959538 Free PMC article. Review. - Temporal and Gene Reassortment Analysis of Influenza C Virus Outbreaks in Hong Kong, SAR, China.
Daniels RS, Galiano M, Ermetal B, Kwong J, Lau CS, Xiang Z, McCauley JW, Lo J. Daniels RS, et al. J Virol. 2022 Feb 9;96(3):e0192821. doi: 10.1128/JVI.01928-21. Epub 2021 Nov 17. J Virol. 2022. PMID: 34787455 Free PMC article. - Identification of One Critical Amino Acid Residue of the Nucleoprotein as a Determinant for In Vitro Replication Fitness of Influenza D Virus.
Yu J, Huang C, Sheng Z, Wang Z, Li F, Wang D. Yu J, et al. J Virol. 2021 Aug 25;95(18):e0097121. doi: 10.1128/JVI.00971-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34190601 Free PMC article. - In situ structure and organization of the influenza C virus surface glycoprotein.
Halldorsson S, Sader K, Turner J, Calder LJ, Rosenthal PB. Halldorsson S, et al. Nat Commun. 2021 Mar 16;12(1):1694. doi: 10.1038/s41467-021-21818-9. Nat Commun. 2021. PMID: 33727554 Free PMC article. - Establishment of a Reverse Genetics System for Influenza D Virus.
Ishida H, Murakami S, Kamiki H, Matsugo H, Takenaka-Uema A, Horimoto T. Ishida H, et al. J Virol. 2020 May 4;94(10):e01767-19. doi: 10.1128/JVI.01767-19. Print 2020 May 4. J Virol. 2020. PMID: 32102883 Free PMC article.
References
- Bourmakina, S. V., and A. García-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. - PubMed
- Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. - PubMed
- Elleman, C. J., and W. S. Barclay. 2004. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 321:144-153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials