Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes - PubMed (original) (raw)
Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes
Benoit Palancade et al. Mol Biol Cell. 2007 Aug.
Abstract
Increasing evidences suggest that nuclear pore complexes (NPCs) control different aspects of nuclear metabolism, including transcription, nuclear organization, and DNA repair. We previously established that the Nup84 complex, a major NPC building block, is part of a genetic network involved in DNA repair. Here, we show that double-strand break (DSB) appearance is linked to a shared function of the Nup84 and the Nup60/Mlp1-2 complexes. Mutants within these complexes exhibit similar genetic interactions and alteration in DNA repair processes as mutants of the SUMO-protease Ulp1. Consistently, these nucleoporins are required for maintenance of proper Ulp1 levels at NPCs and for the establishment of the appropriate sumoylation of several cellular proteins, including the DNA repair factor Yku70. Moreover, restoration of nuclear envelope-associated Ulp1 in nucleoporin mutants reestablishes proper sumoylation patterns and suppresses DSB accumulation and genetic interactions with DNA repair genes. Our results thus provide a molecular mechanism that underlies the connection between NPC and genome stability.
Figures
Figure 1.
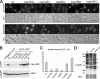
DNA repair foci analysis in nuclear pore and nucleocytoplasmic transport mutants. (A) a, fluorescence microscopy analysis of Rad52-YFP–expressing strains grown at 30°C. The DIC images are also shown. b, quantification of Rad52–YFP foci occurrence for each phase of the cell cycle. c, quantification of the number of Rad52–YFP foci per nucleus. The results of one representative experiment among three is shown, where at least 300 cells were counted for each strain. *, apparent increase in the number of foci in kap121-34 and Δ_N338-ulp1 nup133_Δ mutants is caused by accumulation of cells in G2/M (∼60% in both mutants compared with 25–30% in wt cells). (B) Hyperrecombination assay in nucleoporins, ulp1, and _yku70_Δ mutants. Recombination frequencies were calculated for the indicated strains as described in Materials and Methods.
Figure 2.
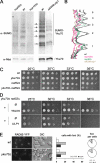
The Nup84 complex and Nup60 are required for Ulp1 localization and stabilization and for the establishment of proper sumoylation patterns. (A) Fluorescence microscopy analysis of ULP1-GFP and NUP49-GFP localization in wild-type and nucleoporin mutant strains. The DIC images are also shown. Cells were grown at 30°C, except for the nup159-1 mutant, which was grown at 22°C to visualize NPC aggregation (Belgareh et al., 1998). (B) Whole cell lysates from _ULP1-GFP_–tagged strains were analyzed by Western blot by using an anti-GFP antibody and an anti-NOP1 (nucleolar protein) antibody as a loading control. The positions of the Ulp1–GFP fusion protein and of Nop1 are indicated. (C) Quantification of the amounts of Ulp1 in the different nucleoporin mutants. Serial dilutions of the samples used in B were analyzed by Western blotting and the integrated intensities of the Ulp1–GFP bands were measured using MetaMorph software. The amounts are expressed as a percentage of the wt value. (D) Total sumoylated proteins from wt, nup120_Δ and ΔN338-ulp1 strains were detected using an anti-SUMO antibody. Similar amounts of proteins were loaded for each lane, as indicated by the amido-black staining of the same blot (“stain_”). Molecular weights (kilodaltons) are indicated. SUMO-conjugates that are decreased in the mutants are indicated by stars, and the increased conjugates are indicated by arrows.
Figure 3.
Decreased levels of Ulp1 at NPCs in Nup84 or Nup60/Mlp1–2 complexes mutants are not merely due to defects in Kap121 or Kap95 pathways. (A) Fluorescence microscopy analysis of NUP60-GFP localization in wild-type or nucleoporin mutant strains grown at 30°C. The DIC images are also shown. (B) Fluorescence microscopy analysis of KAP95-GFP (left) or KAP121-GFP (right) localization in wild-type or nucleoporin mutant strains grown at 30°C. (C) Ulp1-GFP expression levels were analyzed by Western blot by using an anti-GFP antibody in wt, _nup133_Δ, _nup60_Δ, or kap121-34 mutant strains grown at 30°C or shifted for 3 h at 37°C. The anti-NOP1 antibody was used as a loading control. (D) Fluorescence microscopy analysis of wild-type or nucleoporin mutant strains expressing NIC96-mRFP and transformed with the pNOP1-GFP-ULP1 construct. DIC images are also shown.
Figure 4.
Complementation of nucleoporin mutant phenotypes by restoration of nuclear envelope-associated ULP1. (A) Total sumoylated proteins were detected using an anti-SUMO antibody in whole cell extracts from wt, _nup120_Δ, or _nup60_Δ strains that were transformed with the pRS315 (Ø) or the pRS315-NOP1prom-GFP-ULP1 (ULP1) plasmids and treated (+), or not (−), for 2 h with 0.3% MMS. Similar amounts of proteins were loaded for each lane, as indicated by the amido-black staining (“stain”) of the same blot. Molecular weights (kilodaltons) are indicated. Stars indicate SUMO-conjugates, which were decreased in the mutants and have been restored upon ULP1 expression, whereas arrows indicate the conjugates, which were increased in the mutants and reduced upon ULP1 expression. (B) wt, _nup133Δrad52_Δ, and _nup60Δrad52_Δ mutant strains were transformed with pRS315 (Ø) or pRS315 derivatives expressing the indicated constructs under the control of the NOP1 promoter (schematized on the left). Transformants were spotted as fivefold dilutions on synthetic medium lacking leucine and plates were incubated at 25, 28, 30, or 36°C. *, _nup133Δ rad52_Δ strain is not viable in the presence of the GFP-ulp1[C580S] construct. (C) Rad52–YFP foci analysis in wt, _nup133_Δ, and _nup60_Δ cells transformed with either pRS315 (Ø) or pRS315-NOP1prom-GFP-ULP1 (ULP1). The percentage of cells, which show one, two, or three or more Rad52–YFP foci per nucleus, is indicated. The SD corresponds to the sum of the standard deviations for each of the three subcategories.
Figure 5.
DNA repair efficiencies in nucleoporin and ulp1 mutants. (A) Schematic figure that summarizes the principle of the assay is shown. DR, direct repeats, mediating homologous recombination. (B and C) Absolute SSA and NHEJ values were calculated as described in Materials and Methods, and they represent the number of colonies grown in galactose-relative to those grown in glucose-containing medium. The results of three independent experiments are shown along with the corresponding SD, and they are normalized to 100 for the wt value.
Figure 6.
Yku70 sumoylation is altered in _nup60_Δ, _nup120_Δ and ΔN338-ulp1 strains. (A) Exponentially growing wt, _nup60_Δ, _nup120_Δ, and ΔN338-ulp1 cells expressing Myc-tagged Yku70 at its chromosomal locus were treated for 2 h with 0.3% MMS to facilitate the detection of sumoylated forms of Yku70, as reported previously (Zhao and Blobel, 2005). Unmodified and sumoylated Yku70-Myc were immunoprecipitated under denaturing conditions by using an anti-Myc antibody, and they were detected by Western blot analysis with the anti-Myc (bottom) or anti-SUMO (top) antibodies, respectively. The sumoylated forms of Yku70-Myc represent a very minor fraction of the whole Yku70–Myc population, and they were therefore not detected by the anti-Myc antibody. Control experiments using strains bearing either untagged Yku70 or another myc-tagged open reading frame are provided in Supplemental Figure 5. (B) Line-scan analysis of the sumoylated Yku70 patterns in wt, _nup120_Δ, _nup60_Δ (2 independent experiments for these 3 strains), and ΔN338-ulp1 cells were obtained using the MetaMorph software. For each sample, values were normalized to the total amount of immuno-precipitated Yku70 quantified from the anti-Myc Western blot. Numbers (1–5) on the right correspond to the position of the sumoylated bands indicated in A. (C) Segregants of a yku70Δ/+ rad52Δ/+ diploid were spotted as fivefold dilutions on YPD plates, and then they were analyzed for growth at the indicated temperatures. (D) The _yku70Δ rad52_Δ double mutant strain was transformed with pRS313 (Ø), pRS313-YKU70 (YKU70), pRS315 (Ø), or pRS315-NOP1prom-GFP-ULP1 (ULP1). Transformants were spotted as fivefold dilutions on synthetic medium lacking histidine (top) or leucine (bottom), and growth was analyzed at the indicated temperatures. Growth defects of the double mutant are not complemented by ULP1 overexpression, a result consistent with our model in which Yku70 acts downstream of Ulp1 in the DNA repair processes. (E) Rad52 foci analysis in _yku70_Δ cells. Left, Fluorescence microscopy analysis of Rad52-YFP–expressing strains grown at 30°C. The DIC images are also shown. Middle, quantification of Rad52–YFP foci occurrence for each phase of the cell cycle. Right, quantification of the number of Rad52–YFP foci per nucleus. The results of one representative _yku70_Δ strain among four is shown; >150 cells were counted.
Similar articles
- Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality.
Zhao X, Wu CY, Blobel G. Zhao X, et al. J Cell Biol. 2004 Nov 22;167(4):605-11. doi: 10.1083/jcb.200405168. J Cell Biol. 2004. PMID: 15557117 Free PMC article. - Ulp1 association with nuclear pore complexes is required for the maintenance of global SUMOylation.
Ptak C, Saik NO, Wozniak RW. Ptak C, et al. Mol Biol Cell. 2025 Jul 1;36(7):ar81. doi: 10.1091/mbc.E24-12-0563. Epub 2025 May 6. Mol Biol Cell. 2025. PMID: 40327319 - A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance.
Lewis A, Felberbaum R, Hochstrasser M. Lewis A, et al. J Cell Biol. 2007 Aug 27;178(5):813-27. doi: 10.1083/jcb.200702154. J Cell Biol. 2007. PMID: 17724121 Free PMC article. - The dynamics of karyopherin-mediated nuclear transport.
Marelli M, Dilworth DJ, Wozniak RW, Aitchison JD. Marelli M, et al. Biochem Cell Biol. 2001;79(5):603-12. Biochem Cell Biol. 2001. PMID: 11716302 Review. - Regulation of rDNA stability by sumoylation.
Eckert-Boulet N, Lisby M. Eckert-Boulet N, et al. DNA Repair (Amst). 2009 Apr 5;8(4):507-16. doi: 10.1016/j.dnarep.2009.01.015. Epub 2009 Mar 3. DNA Repair (Amst). 2009. PMID: 19261548 Review.
Cited by
- Nuclear pore components affect distinct stages of intron-containing gene expression.
Bonnet A, Bretes H, Palancade B. Bonnet A, et al. Nucleic Acids Res. 2015 Apr 30;43(8):4249-61. doi: 10.1093/nar/gkv280. Epub 2015 Apr 6. Nucleic Acids Res. 2015. PMID: 25845599 Free PMC article. - The yeast nuclear pore complex and transport through it.
Aitchison JD, Rout MP. Aitchison JD, et al. Genetics. 2012 Mar;190(3):855-83. doi: 10.1534/genetics.111.127803. Genetics. 2012. PMID: 22419078 Free PMC article. - SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes.
Zhao X. Zhao X. Mol Cell. 2018 Aug 2;71(3):409-418. doi: 10.1016/j.molcel.2018.07.027. Mol Cell. 2018. PMID: 30075142 Free PMC article. Review. - SUMO orchestrates multiple alternative DNA-protein crosslink repair pathways.
Serbyn N, Bagdiul I, Noireterre A, Michel AH, Suhandynata RT, Zhou H, Kornmann B, Stutz F. Serbyn N, et al. Cell Rep. 2021 Nov 23;37(8):110034. doi: 10.1016/j.celrep.2021.110034. Cell Rep. 2021. PMID: 34818558 Free PMC article. - Evolutionarily conserved genetic interactions with budding and fission yeast MutS identify orthologous relationships in mismatch repair-deficient cancer cells.
Tosti E, Katakowski JA, Schaetzlein S, Kim HS, Ryan CJ, Shales M, Roguev A, Krogan NJ, Palliser D, Keogh MC, Edelmann W. Tosti E, et al. Genome Med. 2014 Sep 17;6(9):68. doi: 10.1186/s13073-014-0068-4. eCollection 2014. Genome Med. 2014. PMID: 25302077 Free PMC article.
References
- Cabal G. G. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases