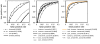RNAstrand: reading direction of structured RNAs in multiple sequence alignments - PubMed (original) (raw)
RNAstrand: reading direction of structured RNAs in multiple sequence alignments
Kristin Reiche et al. Algorithms Mol Biol. 2007.
Abstract
Motivation: Genome-wide screens for structured ncRNA genes in mammals, urochordates, and nematodes have predicted thousands of putative ncRNA genes and other structured RNA motifs. A prerequisite for their functional annotation is to determine the reading direction with high precision.
Results: While folding energies of an RNA and its reverse complement are similar, the differences are sufficient at least in conjunction with substitution patterns to discriminate between structured RNAs and their complements. We present here a support vector machine that reliably classifies the reading direction of a structured RNA from a multiple sequence alignment and provides a considerable improvement in classification accuracy over previous approaches.
Software: RNAstrand is freely available as a stand-alone tool from http://www.bioinf.uni-leipzig.de/Software/RNAstrand and is also included in the latest release of RNAz, a part of the Vienna RNA Package.
Figures
Figure 1
Receiver operating characteristic of all descriptor combinations. Receiver operating characteristic (ROC) for all descriptor combinations. Corresponding AUC is given in brackets. ROC curves were computed by a 5-fold cross-validation on the training data set using plotroc.py of the libsvm 2.8 package [18] after an optimal SVM parameter set was chosen by grid.py. True positive and false positive rates are calculated by interpreting the SVM decision values. Prediction accuracies as plotted here are larger compared to accuracies in Table 1 as even though cross-validation ensures that training and testing is done on different alignments some sequences may occur in the training as well as in the test alignments. In contrast, accuracies in Table 1 are based on test alignments which do not contain any sequence attending at a training alignment.
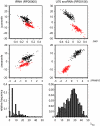
Figure 2
GU base pair dependency. Scatter plots depicting separability between both strands depending on GU base pair content (histograms). Red data points denote alignments in the reading direction of the ncRNA, while black data points belong to their realigned reverse complements. Alignments of tRNAs and U70 snoRNAs do not have significantly different number of sequences nor differ significantly in mean pairwise identity (see Additional file 1). That alignments in reading direction of U70 snoRNA are well separated from their reverse complements compared to alignments containing tRNAs is due to high content of GU base pairs in the secondary structure of U70 snoRNAs.
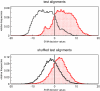
Figure 3
Histogram of SVM decision values. Distribution of SVM decision values of RNAz-positive alignments. The upper histogram belongs to all alignments of the test set. Whereas the lower one shows the distribution of the decision values for shuffled alignments. Columns of the test alignments were randomly permuted to create shuffled alignments. Red dotted bins denote alignments where the ncRNA has the same reading direction as the alignment. Black bins belong to alignments where the ncRNA is contained in the reverse complement. Note that the shuffling procedure does not completely destroy the direction information.
Figure 4
Receiver operating characteristic of all descriptor combinations for shuffled alignments. ROC curves of all descriptor combinations for shuffled alignments. Columns of test alignments were randomly permuted to create shuffled alignments. Corresponding AUC is given in brackets. ROC curves were computed by training a SVM model for each descriptor combination and testing the model on shuffled alignments by utilizing plotroc.py of the libsvm 2.8 package [18]. Training was done with the original training set for RNAstrand. SVM parameter and kernel did not change, i.e. a radial basis function kernel with parameters C = 128 and γ = 0.5 were used.
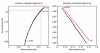
Figure 5
Receiver operating characteristic of test alignments. False positive rates of RNAz-positive test alignments versus true positive rates at different cutoff levels c. The left plot depicts rates in case undecided alignments are included in the calculation. Meaning that the true positive rate is defined as
tptp+fn+u MathType@MTEF@5@5@+=feaafiart1ev1aaatCvAUfKttLearuWrP9MDH5MBPbIqV92AaeXatLxBI9gBaebbnrfifHhDYfgasaacH8akY=wiFfYdH8Gipec8Eeeu0xXdbba9frFj0=OqFfea0dXdd9vqai=hGuQ8kuc9pgc9s8qqaq=dirpe0xb9q8qiLsFr0=vr0=vr0dc8meaabaqaciaacaGaaeqabaqabeGadaaakeaadaWcaaqaaiabdsha0jabdchaWbqaaiabdsha0jabdchaWjabgUcaRiabdAgaMjabd6gaUjabgUcaRiabdwha1baaaaa@3861@
, where tp denotes alignments which have been correctly classified to contain the ncRNA in the same reading direction as the input alignment. fn is the number of alignments which have been falsely classified to contain the ncRNA on the reverse complement, while u contains all alignments which contain the ncRNA in the same reading direction but RNAstrand were not able to predict a reading direction. False positive rate is defined respectively. The right handed plot discards unclassified alignments. Hence, the true positive rate is defined as
tptp+fn MathType@MTEF@5@5@+=feaafiart1ev1aaatCvAUfKttLearuWrP9MDH5MBPbIqV92AaeXatLxBI9gBaebbnrfifHhDYfgasaacH8akY=wiFfYdH8Gipec8Eeeu0xXdbba9frFj0=OqFfea0dXdd9vqai=hGuQ8kuc9pgc9s8qqaq=dirpe0xb9q8qiLsFr0=vr0=vr0dc8meaabaqaciaacaGaaeqabaqabeGadaaakeaadaWcaaqaaiabdsha0jabdchaWbqaaiabdsha0jabdchaWjabgUcaRiabdAgaMjabd6gaUbaaaaa@360C@
and the false positive rate as
fpfp+tn MathType@MTEF@5@5@+=feaafiart1ev1aaatCvAUfKttLearuWrP9MDH5MBPbIqV92AaeXatLxBI9gBaebbnrfifHhDYfgasaacH8akY=wiFfYdH8Gipec8Eeeu0xXdbba9frFj0=OqFfea0dXdd9vqai=hGuQ8kuc9pgc9s8qqaq=dirpe0xb9q8qiLsFr0=vr0=vr0dc8meaabaqaciaacaGaaeqabaqabeGadaaakeaadaWcaaqaaiabdAgaMjabdchaWbqaaiabdAgaMjabdchaWjabgUcaRiabdsha0jabd6gaUbaaaaa@35F0@
. The curves for both SVM decision classes are given. Red curves denote alignments containing the ncRNA in the reading direction of the input alignment. Black curves belong to alignments which contain the ncRNA on the reverse complementary strand. The values of c range from 0 to 0.95 in steps of 0.05.
Similar articles
- Hairpins in a Haystack: recognizing microRNA precursors in comparative genomics data.
Hertel J, Stadler PF. Hertel J, et al. Bioinformatics. 2006 Jul 15;22(14):e197-202. doi: 10.1093/bioinformatics/btl257. Bioinformatics. 2006. PMID: 16873472 - NcDNAlign: plausible multiple alignments of non-protein-coding genomic sequences.
Rose D, Hertel J, Reiche K, Stadler PF, Hackermüller J. Rose D, et al. Genomics. 2008 Jul;92(1):65-74. doi: 10.1016/j.ygeno.2008.04.003. Epub 2008 Jun 3. Genomics. 2008. PMID: 18511233 - LocARNA-P: accurate boundary prediction and improved detection of structural RNAs.
Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R. Will S, et al. RNA. 2012 May;18(5):900-14. doi: 10.1261/rna.029041.111. Epub 2012 Mar 26. RNA. 2012. PMID: 22450757 Free PMC article. - Sparse RNA folding revisited: space-efficient minimum free energy structure prediction.
Will S, Jabbari H. Will S, et al. Algorithms Mol Biol. 2016 Apr 23;11:7. doi: 10.1186/s13015-016-0071-y. eCollection 2016. Algorithms Mol Biol. 2016. PMID: 27110275 Free PMC article. Review. - De novo discovery of structured ncRNA motifs in genomic sequences.
Ruzzo WL, Gorodkin J. Ruzzo WL, et al. Methods Mol Biol. 2014;1097:303-18. doi: 10.1007/978-1-62703-709-9_15. Methods Mol Biol. 2014. PMID: 24639166 Review.
Cited by
- GraphClust2: Annotation and discovery of structured RNAs with scalable and accessible integrative clustering.
Miladi M, Sokhoyan E, Houwaart T, Heyne S, Costa F, Grüning B, Backofen R. Miladi M, et al. Gigascience. 2019 Dec 1;8(12):giz150. doi: 10.1093/gigascience/giz150. Gigascience. 2019. PMID: 31808801 Free PMC article. - Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA.
Moss WN, Steitz JA. Moss WN, et al. BMC Genomics. 2013 Aug 9;14:543. doi: 10.1186/1471-2164-14-543. BMC Genomics. 2013. PMID: 23937650 Free PMC article. - Identification and characterization of novel conserved RNA structures in Drosophila.
Kirsch R, Seemann SE, Ruzzo WL, Cohen SM, Stadler PF, Gorodkin J. Kirsch R, et al. BMC Genomics. 2018 Dec 11;19(1):899. doi: 10.1186/s12864-018-5234-4. BMC Genomics. 2018. PMID: 30537930 Free PMC article. - Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering.
Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Will S, et al. PLoS Comput Biol. 2007 Apr 13;3(4):e65. doi: 10.1371/journal.pcbi.0030065. Epub 2007 Feb 22. PLoS Comput Biol. 2007. PMID: 17432929 Free PMC article. - ViennaRNA Package 2.0.
Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. Lorenz R, et al. Algorithms Mol Biol. 2011 Nov 24;6:26. doi: 10.1186/1748-7188-6-26. Algorithms Mol Biol. 2011. PMID: 22115189 Free PMC article.
References
- Washietl S, Pedersen JS, Korbel JO, Gruber A, Hackermüller J, Hertel J, Lindemeyer M, Reiche K, Stocsits C, Tanzer A, Ucla C, Wyss C, Antonarakis SE, Denoeud F, Lagarde J, Drenkow J, Kapranov P, Gingeras TR, Guigó R, Snyder M, Gerstein MB, Reymond A, Hofacker IL, Stadler PF. Structured RNAs in the ENCODE Selected Regions of the Human Genome. Gen Res. 2007. - PMC - PubMed
LinkOut - more resources
Full Text Sources