Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia - PubMed (original) (raw)
. 2007 Jun 1;129(5):879-90.
doi: 10.1016/j.cell.2007.03.043.
Stephan M Tanner, John C Byrd, Elizabeth B Angerman, James D Perko, Shih-Shih Chen, Björn Hackanson, Michael R Grever, David M Lucas, Jennifer J Matkovic, Thomas S Lin, Thomas J Kipps, Fiona Murray, Dennis Weisenburger, Warren Sanger, Jane Lynch, Patrice Watson, Mary Jansen, Yuko Yoshinaga, Richard Rosenquist, Pieter J de Jong, Penny Coggill, Stephan Beck, Henry Lynch, Albert de la Chapelle, Christoph Plass
Affiliations
- PMID: 17540169
- PMCID: PMC4647864
- DOI: 10.1016/j.cell.2007.03.043
Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia
Aparna Raval et al. Cell. 2007.
Abstract
The heritability of B cell chronic lymphocytic leukemia (CLL) is relatively high; however, no predisposing mutation has been convincingly identified. We show that loss or reduced expression of death-associated protein kinase 1 (DAPK1) underlies cases of heritable predisposition to CLL and the majority of sporadic CLL. Epigenetic silencing of DAPK1 by promoter methylation occurs in almost all sporadic CLL cases. Furthermore, we defined a disease haplotype, which segregates with the CLL phenotype in a large family. DAPK1 expression of the CLL allele is downregulated by 75% in germline cells due to increased HOXB7 binding. In the blood cells from affected family members, promoter methylation results in additional loss of DAPK1 expression. Thus, reduced expression of DAPK1 can result from germline predisposition, as well as epigenetic or somatic events causing or contributing to the CLL phenotype.
Figures
Figure 1
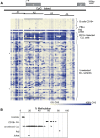
DAPK1 Promoter Methylation in CLL Samples (A) Schematic representation of the DAPK1 gene showing the location of the CpG island (black bar) and the four bisulfite reaction amplicons (A1–A4). The arrow indicates the predicted transcription start site. The lower panel shows a graphical display of quantitative DNA methylation data for the DAPK1 promoter region. Each square represents a single CpG or a group of two or three CpGs analyzed, and each row represents a sample. Methylation frequencies extend from light yellow (0%) to dark blue (100%). Gray indicates unavailable data. Samples included seven CD19+ selected control B cells, four control normal PBMCs, seven CD19+ selected CLL cells, and PBMCs from 62 sporadic CLL samples, Raji and Jurkat cells. Asterisks indicate CLL samples that showed less than 11% methylation. (B) Plot of average percentage methylation in regions A1–A4, in controls (seven CD19+ selected B cells and four PBL samples; total = 11), CD19+ selected CLL cells (n = 7), unselected CLL cells (n = 62), and Raji and Jurkat cell lines.
Figure 2
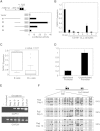
Epigenetic Silencing of DAPK1 in CLL (A) pGL3 luciferase constructs ligated with different DAPK1 5′ upstream inserts (nos. 1–4) were transfected into Jurkat cells, and reporter activity was studied 48 hr after transfection. pGL3 vector without insert was used as a negative control. Error bars indicate standard deviation (SD). (B) Quantitative DAPK1 expression in normal B cells and seven CD19+ selected CLL samples. The expression in CLL samples is shown relative to the expression in B cells (defined as 1.0). Error bars indicate SD. (C) Box plot of DAPK1 expression in 50 CLL samples as measured by semiquantitative RT-PCR and compared to its expression in six normal B cell samples. The distribution is significantly different (p < 0.01). (D) Luciferase assays in 293T cells with either methylated or unmethylated DAPK1 construct no. 2. Error bars indicate SD. (E) DAPK1 expression in Raji cells treated with 0.5 μM decitabine. RT-PCR was performed for DAPK1 and GAPDH on untreated and treated cell lines. Jurkat cells were used as a positive control. (F) Bisulfite sequences in BS1 region (c.1-1509–c.1-1262) for untreated, 6 and 12 days decitabine-treated (0.5 μM) Raji cells. Each row represents a clone. The open circles indicate unmethylated CpG, and closed circles indicate methylated CpG dinucleotides. The overall methylation frequency is given in percentage. Error bars indicate SD.
Figure 3
DAPK1 Regulates Apoptosis in Lymphoid Cells (A) DAPK1 expression in Jurkat cells stably transfected with either vector alone, DAPK1 siRNA-A, or DAPK1 siRNA-C as measured by western blot. Tubulin expression served as a control. (B) Percent live cells were measured in Jurkat cells stably transfected with vector alone or DAPK1 siRNA-C, treated with activating-Fas antibody (100 ng/ml). After 16 hr, cells were harvested and suspended in binding buffer with annexin V-FITC and propidium iodide, followed by flow cytometry to assess cell death. Error bars indicate SD. (C) p53 expression in cells treated with either no, 50 ng/ml, or 100 ng/ml Fas-activating antibody.
Figure 4
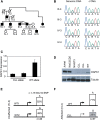
DAPK1 Expression in CLL Family 4532 (A) Pedigree of CLL family no. 4532. Open circle and square represent unaffected female and male, respectively, while closed circle and square represent affected female and male. (B) Sequencing of SNP c.114A > G from genomic DNA and cDNA of two unaffected (IV-3 and IV-4) and two affected (III-3 and III-4) individuals. (C) RT-PCR on RNA isolated from monochromosomal hybrid clones with either WT or the CLL chromosome 9 from fibroblast cells of individual III-4 is shown. RPL4 expression was used as an internal control. Error bars indicate SD. (D) DAPK1 protein expression from WT and CLL alleles in monochromosomal hybrids. Jurkat and WaC3CD5 cells were used as positive controls, and NIH3T3 cells were used as the negative control to show specificity to human DAPK1. (E and F) The RT-PCR product amplifying SNP c.1510A > G in unaffected (IV-3; E) and affected (III-4; F) fibroblast cell lines was cloned, and individual clones were genotyped. Shown is the percentage of A and G clones in IV-3 and III-4, and n is the number of clones studied. The difference in allelic expression of DAPK1 in III-4 and IV-3 was statistically significant (p < 0.01).
Figure 5
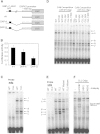
An A to G Change at c.1-6531 bp Regulates DAPK1 Expression in CLL Family (A) A 357 bp PCR product with SNP c.1-6531A > G was ligated upstream to DAPK1 promoter (c.1-2215–c.1-1151) luciferase construct with either A (DAP-A) or G (DAP-G) as SNP. (B) The DAPK1 promoter alone (no. 1), DAP-A, or DAP-G constructs were transfected into Jurkat cells, and the luciferase activity was measured. Error bars indicate SD. (C) Nuclear extracts from Jurkat cells were analyzed by EMSA assay using WT or CLL oligo. Five specific bands (I–V) are marked. (D) Ten- or fifty-fold molar excess concentrations of cold oligos were used for competition assays. An unlabeled Oct-1 oligo was used as a negative control. (E) EMSA assays were performed using WT, WT mutant, CLL, and CLL mutant oligos. In mutant oligos, the adjacent bases to the c.1-6531 SNP were mutated. (F) For the supershift assay, antibodies against HOXB7, USF2, and MSX2 were added to the Jurkat nuclear extract, and gel shift was studied using the CLL oligo.
Figure 6
Downregulation of DAPK1 Expression by HOXB7 (A and B) Fibroblast cell lines from affected (III-4) and unaffected (IV-4) family members were transfected with 60 nM of HOXB7 siRNA or scrambled siRNA, and HOXB7 expression was studied at different time points by semiquantitative SYBR green RT-PCR. HOXB7 expression in untreated cells was set as 1.0. Error bars indicate SD. (C and D) DAPK1 expression was studied in the fibroblast cell lines transfected with HOXB7 siRNA or scrambled siRNA for different time points by quantitative SYBR green RT-PCR. DAPK1 expression in untreated cells was set as 1.0. Error bars indicate SD. (E and F) Allele-specific expression of DAPK1 in an affected (E) and unaffected (F) family member was studied in fibroblasts from III-4 and IV-4, before and after transfection of HOXB7 siRNA. RT-PCR products including SNP c.1510A>G were cloned and genotyped. Shown is the percentage of the number of clones from the two alleles, and (n) indicates number of clones studied.
Figure 7
DAPK1 Expression and Promoter Methylation in CLL Cells of Family No. 4532 (A) RT-PCR for DAPK1 with GAPDH as an internal control on RNA extracted from blood cells of no. 4532 family members III-1, III-2, III-3, III-4 and IV-5 and from selected CD19+ normal B-cells from 3 healthy volunteers. Error bars indicate SD. (B) DNA methylation analysis in CLL cells from CD19+ selected control B-cells, total PBMCs from one unaffected family member (IV-1) and two affected individuals (III-1 and III-4). Bisulfite treated DNA was amplified for the BS1 and BS2 regions. Each row represents a clone. The open circles indicate unmethylated and closed circles indicate methylated CpGs.
Comment in
- Chronic lymphocytic leukemia: keeping cell death at bay.
Debatin KM. Debatin KM. Cell. 2007 Jun 1;129(5):853-5. doi: 10.1016/j.cell.2007.05.023. Cell. 2007. PMID: 17540163
Similar articles
- Germline allele-specific expression of DAPK1 in chronic lymphocytic leukemia.
Wei QX, Claus R, Hielscher T, Mertens D, Raval A, Oakes CC, Tanner SM, de la Chapelle A, Byrd JC, Stilgenbauer S, Plass C. Wei QX, et al. PLoS One. 2013;8(1):e55261. doi: 10.1371/journal.pone.0055261. Epub 2013 Jan 28. PLoS One. 2013. PMID: 23383130 Free PMC article. - Chronic lymphocytic leukemia: keeping cell death at bay.
Debatin KM. Debatin KM. Cell. 2007 Jun 1;129(5):853-5. doi: 10.1016/j.cell.2007.05.023. Cell. 2007. PMID: 17540163 - A noncanonical Flt3ITD/NF-κB signaling pathway represses DAPK1 in acute myeloid leukemia.
Shanmugam R, Gade P, Wilson-Weekes A, Sayar H, Suvannasankha A, Goswami C, Li L, Gupta S, Cardoso AA, Baghdadi TA, Sargent KJ, Cripe LD, Kalvakolanu DV, Boswell HS. Shanmugam R, et al. Clin Cancer Res. 2012 Jan 15;18(2):360-369. doi: 10.1158/1078-0432.CCR-10-3022. Epub 2011 Nov 17. Clin Cancer Res. 2012. PMID: 22096027 Free PMC article. - DNA methylation, epimutations and cancer predisposition.
Dobrovic A, Kristensen LS. Dobrovic A, et al. Int J Biochem Cell Biol. 2009 Jan;41(1):34-9. doi: 10.1016/j.biocel.2008.09.006. Epub 2008 Sep 16. Int J Biochem Cell Biol. 2009. PMID: 18835361 Review. - Molecular profiling of chronic lymphocytic leukaemia: genetics meets epigenetics to identify predisposing genes.
Plass C, Byrd JC, Raval A, Tanner SM, de la Chapelle A. Plass C, et al. Br J Haematol. 2007 Dec;139(5):744-52. doi: 10.1111/j.1365-2141.2007.06875.x. Epub 2007 Oct 24. Br J Haematol. 2007. PMID: 17961188 Review.
Cited by
- Methods for cancer epigenome analysis.
Nagarajan RP, Fouse SD, Bell RJ, Costello JF. Nagarajan RP, et al. Adv Exp Med Biol. 2013;754:313-38. doi: 10.1007/978-1-4419-9967-2_15. Adv Exp Med Biol. 2013. PMID: 22956508 Free PMC article. Review. - Cul3-KLHL20 ubiquitin ligase: physiological functions, stress responses, and disease implications.
Chen HY, Liu CC, Chen RH. Chen HY, et al. Cell Div. 2016 Apr 1;11:5. doi: 10.1186/s13008-016-0017-2. eCollection 2016. Cell Div. 2016. PMID: 27042198 Free PMC article. Review. - Prospecting for Breast Cancer Blood Biomarkers: Death-Associated Protein Kinase 1 (DAPK1) as a Potential Candidate.
Arko-Boham B, Owusu BA, Aryee NA, Blay RM, Owusu EDA, Tagoe EA, Adams AR, Gyasi RK, Adu-Aryee NA, Mahmood S. Arko-Boham B, et al. Dis Markers. 2020 May 24;2020:6848703. doi: 10.1155/2020/6848703. eCollection 2020. Dis Markers. 2020. PMID: 32566040 Free PMC article. - Azacitidine in fludarabine-refractory chronic lymphocytic leukemia: a phase II study.
Malik A, Shoukier M, Garcia-Manero G, Wierda W, Cortes J, Bickel S, Keating MJ, Estrov Z. Malik A, et al. Clin Lymphoma Myeloma Leuk. 2013 Jun;13(3):292-5. doi: 10.1016/j.clml.2012.11.009. Epub 2012 Dec 21. Clin Lymphoma Myeloma Leuk. 2013. PMID: 23265768 Free PMC article. Clinical Trial. - Cullin 3 Ubiquitin Ligases in Cancer Biology: Functions and Therapeutic Implications.
Chen HY, Chen RH. Chen HY, et al. Front Oncol. 2016 May 2;6:113. doi: 10.3389/fonc.2016.00113. eCollection 2016. Front Oncol. 2016. PMID: 27200299 Free PMC article. Review.
References
- Amundadottir L.T., Thorvaldsson S., Gudbjartsson D.F., Sulem P., Kristjansson K., Arnason S., Gulcher J.R., Bjornsson J., Kong A., Thorsteinsdottir U., Stefansson K. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. - PMC - PubMed
- Bialik S., Kimchi A. The Death-Associated Protein Kinases: Structure, Function, and Beyond. Annu. Rev. Biochem. 2006;75:189–200. - PubMed
- Bullrich F., Fujii H., Calin G., Mabuchi H., Negrini M., Pekarsky Y., Rassenti L., Alder H., Reed J.C., Keating M.J. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 2001;61:6640–6648. - PubMed
- Calin G.A., Croce C.M. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin. Oncol. 2006;33:167–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA101956/CA/NCI NIH HHS/United States
- P01 CA101956/CA/NCI NIH HHS/United States
- U01 CA086389/CA/NCI NIH HHS/United States
- CA81534/CA/NCI NIH HHS/United States
- P01 CA081534/CA/NCI NIH HHS/United States
- T32 CA106196/CA/NCI NIH HHS/United States
- R21 CA110496/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- P30 CA16058/CA/NCI NIH HHS/United States
- 5U01 CA86389/CA/NCI NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
- CA110496/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases