Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin - PubMed (original) (raw)
Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin
Sergi Garcia-Manyes et al. Biophys J. 2007.
Abstract
Single-protein force experiments have relied on a molecular fingerprint based on tethering multiple single-protein domains in a polyprotein chain. However, correlations between these domains remain an issue in interpreting force spectroscopy data, particularly during protein folding. Here we first show that force-clamp spectroscopy is a sensitive technique that provides a molecular fingerprint based on the unfolding step size of four single-monomer proteins. We then measure the force-dependent unfolding rate kinetics of ubiquitin and I27 monomers and find a good agreement with the data obtained for the respective polyproteins over a wide range of forces, in support of the Markovian hypothesis. Moreover, with a large statistical ensemble at a single force, we show that ubiquitin monomers also exhibit a broad distribution of unfolding times as a signature of disorder in the folded protein landscape. Furthermore, we readily capture the folding trajectories of monomers that exhibit the same stages in folding observed for polyproteins, thus eliminating the possibility of entropic masking by other unfolded modules in the chain or domain-domain interactions. On average, the time to reach the I27 folded length increases with increasing quenching force at a rate similar to that of the polyproteins. Force-clamp spectroscopy at the single-monomer level reproduces the kinetics of unfolding and refolding measured using polyproteins, which proves that there is no mechanical effect of tethering proteins to one another in the case of ubiquitin and I27.
Figures
FIGURE 1
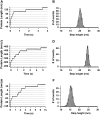
Typical length-versus-time recording of a concatamer of (A) Ubi9, (C) I278, and (E) PoteinL8 stretched at 110 pN, 150 pN, and 80 pN, respectively, yielding staircases in which every single step corresponds to the unfolding of a single module in the polyprotein chain. The measured step height is characteristic of each individual protein, thus serving as a fingerprint of the unfolded protein. Histograms of the measured step heights are shown for Ubi9 pulled at 110 pN (B), I278 pulled at 150 pN (D), and ProteinL8 pulled at 80 pN (F). Gaussian fits to the histograms give rise to step heights of 20.4 ± 0.7 nm (Ubi9), 24.5 ± 0.5 nm (I278), and 15.8 ± 0.9 nm (ProteinL8).
FIGURE 2
Single-protein monomers can be unambiguously unfolded with force-clamp spectroscopy. (A) Scheme of the experimental setup, where the single-protein monomer is held between the cantilever tip and the surface. (B) Five experimental length-versus-time recordings of ubiquitin monomers (red traces) stretched at 110 pN and of I27 monomers (blue traces) stretched at 150 pN, exhibiting well-defined steps at ∼20.0 nm and ∼24.5 nm, respectively. In all curves, an initial step before the unfolding step is observed (red and blue squares), described in the text. From this step size, the length of the folded protein with the handles can be measured for every particular trace, as shown in the histograms in C, (ubiquitin, red; I27, blue), which agrees with a random probability of pickup along the handles. (D) Histogram corresponding to all the recorded extensions for ubiquitin (red; n =1631) and I27 (blue; n =1317). Gaussian fit to the peaks in both histograms give rise to 20.65 ± 0.91 nm (ubiquitin) and 24.50 ± 0.77 nm (I27), as in the polyproteins (Fig. 1).
FIGURE 3
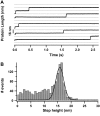
(A) Typical length-versus-time recordings corresponding to the unfolding of a ProteinL monomer when pulled at 80 pN, yielding a unique step with a measured step height of ∼16 nm (discontinuous lines are to guide the eye). (B) Histogram corresponding to all the measured variations in length during four individual experiments at stretching forces ranging from 40 to 100 pN (n = 1499). Gaussian fit to the histogram peak yields a mean step height of 15.3 ± 2.16 nm.
FIGURE 4
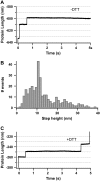
Chemical reactions can be monitored with single-protein monomers. (A) Typical length-versus-time recording showing the unfolding of a single I27G32C-A75C monomer stretched at 130 pN as confirmed by the presence of a unique ∼10.5-nm step. (B) Histogram of all the measured step sizes (n = 329), which shows a preferential peak centered at ∼10.5 nm. When the same experiment is repeated in a reducing environment (50 mM DTT), the length-versus-time traces exhibit two steps (C), the first one (∼10.5 nm) corresponding to the unfolding of the I27G32C-A75C monomer, and the second one (∼13.8 nm) corresponding to the reduction of the solvent-exposed disulfide bond, therefore releasing the previously sequestered amino acids.
FIGURE 5
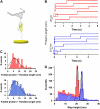
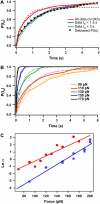
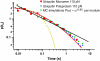
Unfolding kinetics for I27 and ubiquitin monomers present a close agreement with polyprotein data. (A) Unfolding probability of ubiquitin monomer at 110 pN. When all trajectories (n = 343) are included (red line), a single exponential fit (not constrained to 1, dashed red line) to the unfolding probability gives α = 1.53 s−1. The fit (gray dashed line) to the unfolding probability of the trajectories with detachment time greater than 1.5 s (blue line) and 3 s (green line) gives α = 0.9 s−1. Black squares show the cumulative probability of unfolding P(_t_u) obtained from unbiasing all the experimental data (red line) from their experimental detachment rate distribution according to Eq. 1 in the text. A single exponential fit to this curve yields α = 0.9 s−1. (B) P(_t_u) distribution for ubiquitin monomer unfolding at 90, 110, 130, 150, and 170 pN. Distributions at each force are fitted to a single exponential curve (dashed lines) yielding the rate of unfolding at each particular force in a two-state model. (C) Plot of the logarithm of the rate constant as a function of the pulling force for ubiquitin (red) and I27 (blue). In both cases, circles stand for polyproteins and squares for the respective monomers. Data for polyubiquitin rates are extracted from Schlierf et al. (28). PolyI27 rates are obtained as shown in Fig. 7. Linear fit of Eq. 2 in the text to ubiquitin data (red continuous line) yields α_0 = 1.5 × 10−2 s−1 and Δ_x = 0.16 nm and to I27 data (blue continuous line) yields α_0 = 7.2 × 10−4 s−1 and Δ_x = 0.19 nm.
FIGURE 6
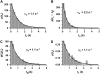
Monte-Carlo simulations validate the method for unbiasing the unfolding and the detachment rate distributions. (A) p(_t_u) distribution corresponding to 1000 independent simulations of ubiquitin monomer pulled at 110 pN assuming a two-state unfolding process without any effect of the detachment rate, giving rise to an unfolding constant α = 1.1 s−1. (B) distribution corresponding to 1000 independent simulations of ubiquitin monomer pulled at 110 pN with the effect of an exponentially distributed detachment rate of _α_d = 1.1 s−1 (distribution shown in C). Only in 511 of the 1000 attempts was the unfolding event measured, resulting in a “biased” unfolding constant α = 2.2 s−1. (D) p(_t_u) distribution corresponding to unbiasing the
distribution shown in B by the associated detachment time distribution
shown in C by using Eq. 1 in the text. The resulting p(_t_u) distribution follows an unfolding kinetics with α = 1.1 s−1, thus recovering the unfolding rate obtained in A when no effect of the detachment time was present.
FIGURE 7
Five averaged and normalized polyI27 unfolding time courses obtained at different stretching forces: 90 pN, 150 pN, 170 pN, 190 pN, and 200 pN. Discontinuous black lines correspond to single exponential fits with rate constants presented as blue squares in Fig. 5 C in the text.
FIGURE 8
p(_t_u) of ubiquitin monomer (red squares) and polyprotein (blue squares) after logarithmic binning. The distribution of unfolding times is fitted to a log normal distribution (solid red line) and to a power law (solid black line), both deviating from the single exponential fit (yellow dashed line). Green triangles stand for the generated p(_t_u) that corresponds to the predicted P(α) ≈ _t_−0.85 for a single monomer in Monte Carlo simulations (31). All the distributions are normalized by the area under the curve.
FIGURE 9
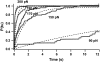
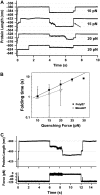
Folding times of single I27 monomers depend on the quenching force. (A) Experimental folding trajectories of the I27 monomer. In all cases, a two-pulse procedure has been used. The monomer is stretched at 120 pN for 4 s to unfold the protein, as revealed by the ∼24.5-nm step (fingerprint of I27 unfolding) after the initial elastic elongation step (see Fig. 2 C). Subsequently, the force is quenched to a lower force (10 pN, 15 pN, or 20 pN) to trigger folding for 3 s. Finally, the force is again increased to 120 pN to unfold the monomer for another 3 s. In the upper trajectory the quenching force was set to 10 pN, and the folding process was almost instantaneous, thus showing an apparent two-state behavior. When the quenching force was set to 15 pN, the folding time was increased to ∼0.8 s, thus resulting in a more complex folding trajectory. Finally, when the quenching force was set to 20 pN, the folding time increased to ∼2.2 s or no folding was observed (failure trajectory). After the collapse takes place, the protein is stretched at higher force to promote the monomer reunfolding, marked by the steplike increase in length of ∼24.5 nm. In the second trajectory (15 pN), the step occurs very fast, as shown by an arrow. (B) Logarithmic plot of the folding time, τ, as a function of the quenching force, for the polyprotein (black squares) and the monomer (white circles) I27. Data were fitted to an exponential relationship, yielding _τ_F = 0.52 exp (F × 0.1) for the polyI27 (black solid line) and _τ_F = 0.35 exp (F × 0.09) for the I27 monomer (gray solid line). (C) Typical force quench trajectory of ubiquitin. The monomer is pulled at 110 pN, and once it unfolds, the force is quenched down to 15 pN for 6 s. Within this period of time, the protein exhibits a contraction in length that leads to the folded length. Subsequently, the force is raised again to 110 pN to reunfold the monomer. The unfolding events can be easily identified in the force trace as “spikes” corresponding to the force feedback time.
FIGURE 10
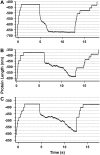
(A) Three typical folding trajectories of an I278 polyprotein. The polyprotein was unfolded at high force (150 pN) as confirmed by the stepwise ∼24.5-nm increment in length. Then, the force is quenched down to (A) 15 pN, (B) 20 pN, or (C) 25 pN so as to favor folding. As has been previously described for polyubiquitin (33), folding trajectories, unlike unfolding ones, occur cooperatively and feature large fluctuations in length during the pathway that leads the protein from the unfolded state to the folded state. Therefore, the general folding trends observed for polyubiquitin trajectories also apply to polyI27. Note that, on average, the lower the quenching force, the higher the time the molecule needs to fold (A–C).
Similar articles
- Dwell-time distribution analysis of polyprotein unfolding using force-clamp spectroscopy.
Brujic J, Hermans RI, Garcia-Manyes S, Walther KA, Fernandez JM. Brujic J, et al. Biophys J. 2007 Apr 15;92(8):2896-903. doi: 10.1529/biophysj.106.099481. Epub 2007 Jan 26. Biophys J. 2007. PMID: 17259284 Free PMC article. - Direct observation of markovian behavior of the mechanical unfolding of individual proteins.
Cao Y, Kuske R, Li H. Cao Y, et al. Biophys J. 2008 Jul;95(2):782-8. doi: 10.1529/biophysj.107.128298. Epub 2008 Mar 28. Biophys J. 2008. PMID: 18375518 Free PMC article. - The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques.
Schlierf M, Li H, Fernandez JM. Schlierf M, et al. Proc Natl Acad Sci U S A. 2004 May 11;101(19):7299-304. doi: 10.1073/pnas.0400033101. Epub 2004 Apr 27. Proc Natl Acad Sci U S A. 2004. PMID: 15123816 Free PMC article. - Single molecule force spectroscopy using polyproteins.
Hoffmann T, Dougan L. Hoffmann T, et al. Chem Soc Rev. 2012 Jul 21;41(14):4781-96. doi: 10.1039/c2cs35033e. Epub 2012 May 30. Chem Soc Rev. 2012. PMID: 22648310 Review. - Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering.
Carrion-Vazquez M, Oberhauser AF, Fisher TE, Marszalek PE, Li H, Fernandez JM. Carrion-Vazquez M, et al. Prog Biophys Mol Biol. 2000;74(1-2):63-91. doi: 10.1016/s0079-6107(00)00017-1. Prog Biophys Mol Biol. 2000. PMID: 11106807 Review.
Cited by
- Unequivocal single-molecule force spectroscopy of proteins by AFM using pFS vectors.
Oroz J, Hervás R, Carrión-Vázquez M. Oroz J, et al. Biophys J. 2012 Feb 8;102(3):682-90. doi: 10.1016/j.bpj.2011.12.019. Epub 2012 Feb 7. Biophys J. 2012. PMID: 22325292 Free PMC article. - Force-activated reactivity switch in a bimolecular chemical reaction.
Garcia-Manyes S, Liang J, Szoszkiewicz R, Kuo TL, Fernández JM. Garcia-Manyes S, et al. Nat Chem. 2009 Jun;1(3):236-42. doi: 10.1038/nchem.207. Epub 2009 May 10. Nat Chem. 2009. PMID: 21378854 - Probing DNA base pairing energy profiles using a nanopore.
Viasnoff V, Chiaruttini N, Bockelmann U. Viasnoff V, et al. Eur Biophys J. 2009 Feb;38(2):263-9. doi: 10.1007/s00249-008-0372-2. Epub 2008 Oct 3. Eur Biophys J. 2009. PMID: 18836709 - Titin domains progressively unfolded by force are homogenously distributed along the molecule.
Bianco P, Mártonfalvi Z, Naftz K, Kőszegi D, Kellermayer M. Bianco P, et al. Biophys J. 2015 Jul 21;109(2):340-5. doi: 10.1016/j.bpj.2015.06.002. Biophys J. 2015. PMID: 26200869 Free PMC article. - FliI6-FliJ molecular motor assists with unfolding in the type III secretion export apparatus.
Kucera J, Terentjev EM. Kucera J, et al. Sci Rep. 2020 Apr 28;10(1):7127. doi: 10.1038/s41598-020-63330-y. Sci Rep. 2020. PMID: 32346005 Free PMC article.
References
- Dietz, H., M. Bertz, M. Schlierf, F. Berkemeier, T. Bornschlogl, J. P. Junker, and M. Rief. 2006. Cysteine engineering of polyproteins for single-molecule force spectroscopy. Nat. Protocols. 1:80–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources