Time variations in the transmissibility of pandemic influenza in Prussia, Germany, from 1918-19 - PubMed (original) (raw)
Time variations in the transmissibility of pandemic influenza in Prussia, Germany, from 1918-19
Hiroshi Nishiura. Theor Biol Med Model. 2007.
Abstract
Background: Time variations in transmission potential have rarely been examined with regard to pandemic influenza. This paper reanalyzes the temporal distribution of pandemic influenza in Prussia, Germany, from 1918-19 using the daily numbers of deaths, which totaled 8911 from 29 September 1918 to 1 February 1919, and the distribution of the time delay from onset to death in order to estimate the effective reproduction number, Rt, defined as the actual average number of secondary cases per primary case at a given time.
Results: A discrete-time branching process was applied to back-calculated incidence data, assuming three different serial intervals (i.e. 1, 3 and 5 days). The estimated reproduction numbers exhibited a clear association between the estimates and choice of serial interval; i.e. the longer the assumed serial interval, the higher the reproduction number. Moreover, the estimated reproduction numbers did not decline monotonically with time, indicating that the patterns of secondary transmission varied with time. These tendencies are consistent with the differences in estimates of the reproduction number of pandemic influenza in recent studies; high estimates probably originate from a long serial interval and a model assumption about transmission rate that takes no account of time variation and is applied to the entire epidemic curve.
Conclusion: The present findings suggest that in order to offer robust assessments it is critically important to clarify in detail the natural history of a disease (e.g. including the serial interval) as well as heterogeneous patterns of transmission. In addition, given that human contact behavior probably influences transmissibility, individual countermeasures (e.g. household quarantine and mask-wearing) need to be explored to construct effective non-pharmaceutical interventions.
Figures
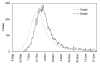
Figure 1
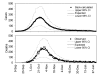
Epidemic curve of pandemic influenza in Prussia, Germany, from 1918–19. Reported daily number of influenza deaths (solid line) and the back-calculated temporal distribution of onset cases (dashed line). Daily counts of onset cases were obtained using the time delay distribution from onset to death (see Table 1). Data source: ref [18] (see [Additional file 1]).
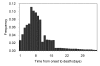
Figure 2
Distribution of the time delay from onset to death during the influenza epidemic in Prussia, Germany, from 1918–19. Time from disease onset (i.e. fever) to death is given for 6233 influenza deaths. A simple 5-day moving average was applied to the original data. Data source: ref [18] (see [Additional file 2]).
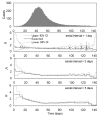
Figure 3
Epidemic curve and the corresponding effective reproduction numbers (R) with variable serial intervals. Time variation in the effective reproduction number (the number of secondary infections generated per case by generation) assuming three different serial intervals is shown. The serial interval was assumed to be 1 (second from the top), 3 (lower middle) and 5 days (bottom). Days are counted from September 9, 1918, onwards.
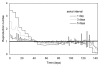
Figure 4
Comparison of the effective reproduction number assuming different serial intervals. Expected values of the effective reproduction number with a serial interval of 1 (grey), 3 (dashed black) and 5 days (solid black). The horizontal solid line represents the threshold value, R = 1, below which the epidemic will decline to extinction. Days are counted from September 9, 1918, onwards.
Figure 5
Simulated epidemic curve of pandemic influenza in Prussia, Germany, from 1918–19. Comparison of observed epidemic curves of onset (top) and death (bottom) with simulated curves. Expected values of influenza cases and deaths (solid line) mainly overlapped with the observed numbers (dot). Dashed lines indicate the corresponding upper and lower 95% confidence intervals (CI) based on 1000 simulation runs. The 95% CI of cases and deaths were determined by 2.5th and 97.5th percentiles of the simulated cases and deaths at each time point.
Similar articles
- Household and community transmission of the Asian influenza A (H2N2) and influenza B viruses in 1957 and 1961.
Nishiura H, Chowell G. Nishiura H, et al. Southeast Asian J Trop Med Public Health. 2007 Nov;38(6):1075-83. Southeast Asian J Trop Med Public Health. 2007. PMID: 18613549 - The 1918-1919 influenza pandemic in England and Wales: spatial patterns in transmissibility and mortality impact.
Chowell G, Bettencourt LM, Johnson N, Alonso WJ, Viboud C. Chowell G, et al. Proc Biol Sci. 2008 Mar 7;275(1634):501-9. doi: 10.1098/rspb.2007.1477. Proc Biol Sci. 2008. PMID: 18156123 Free PMC article. - Comparative estimation of the reproduction number for pandemic influenza from daily case notification data.
Chowell G, Nishiura H, Bettencourt LM. Chowell G, et al. J R Soc Interface. 2007 Feb 22;4(12):155-66. doi: 10.1098/rsif.2006.0161. J R Soc Interface. 2007. PMID: 17254982 Free PMC article. - Estimates of the reproduction numbers of Spanish influenza using morbidity data.
Vynnycky E, Trindall A, Mangtani P. Vynnycky E, et al. Int J Epidemiol. 2007 Aug;36(4):881-9. doi: 10.1093/ije/dym071. Epub 2007 May 21. Int J Epidemiol. 2007. PMID: 17517812 - Transmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic.
Viboud C, Tam T, Fleming D, Handel A, Miller MA, Simonsen L. Viboud C, et al. Vaccine. 2006 Nov 10;24(44-46):6701-7. doi: 10.1016/j.vaccine.2006.05.067. Epub 2006 Jun 9. Vaccine. 2006. PMID: 16806596
Cited by
- The R0 package: a toolbox to estimate reproduction numbers for epidemic outbreaks.
Obadia T, Haneef R, Boëlle PY. Obadia T, et al. BMC Med Inform Decis Mak. 2012 Dec 18;12:147. doi: 10.1186/1472-6947-12-147. BMC Med Inform Decis Mak. 2012. PMID: 23249562 Free PMC article. - Statistical Estimation of the Reproductive Number From Case Notification Data.
White LF, Moser CB, Thompson RN, Pagano M. White LF, et al. Am J Epidemiol. 2021 Apr 6;190(4):611-620. doi: 10.1093/aje/kwaa211. Am J Epidemiol. 2021. PMID: 33034345 Free PMC article. Review. - A biological model for influenza transmission: pandemic planning implications of asymptomatic infection and immunity.
Mathews JD, McCaw CT, McVernon J, McBryde ES, McCaw JM. Mathews JD, et al. PLoS One. 2007 Nov 28;2(11):e1220. doi: 10.1371/journal.pone.0001220. PLoS One. 2007. PMID: 18043733 Free PMC article. - Tracking Epidemics With Google Flu Trends Data and a State-Space SEIR Model.
Dukic V, Lopes HF, Polson NG. Dukic V, et al. J Am Stat Assoc. 2012;107(500):1410-1426. doi: 10.1080/01621459.2012.713876. Epub 2012 Dec 21. J Am Stat Assoc. 2012. PMID: 37583443 Free PMC article. - Aspectos básicos de la transmisibilidad.
Gil Cuesta J, Vaqué Rafart J. Gil Cuesta J, et al. Vacunas. 2008 Jan;9(1):25-33. doi: 10.1016/S1576-9887(08)71918-6. Epub 2009 Jan 6. Vacunas. 2008. PMID: 32288705 Free PMC article. Review. Spanish. No abstract available.
References
- Dietz K. The estimation of the basic reproduction number for infectious diseases. Stat Methods Med Res. 1993;2:23–41. - PubMed
- Smith CE. Factors in the transmission of virus infections from animal to man. Sci Basis Med Annu Rev. 1964:125–150. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical