Molecular dynamics using atomic-resolution structure reveal structural fluctuations that may lead to polymerization of human Cu-Zn superoxide dismutase - PubMed (original) (raw)
Molecular dynamics using atomic-resolution structure reveal structural fluctuations that may lead to polymerization of human Cu-Zn superoxide dismutase
Richard W Strange et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Mutations of the gene encoding Cu-Zn superoxide dismutase (SOD1) cause 20% of the familial cases of the progressive neurodegenerative disease ALS. A growing body of evidence suggests that in familial ALS (FALS) it is the molecular behavior of the metal-depleted SOD1 dimer that leads to a gain of toxic properties by misfolding, unfolding, and aggregation. Structural studies have so far provided static snapshots on the behavior of the wild-type enzyme and some of the FALS mutants. New approaches are required to map out the structural trajectories of the molecule. Here, using our 1.15-A resolution structure of fully metallated human SOD1 and highly parallelized molecular dynamics code on a high-performance capability computer, we have undertaken molecular dynamics calculations to 4,000 ps to reveal the first stages of misfolding caused by metal deletion. Large spatial and temporal fluctuations of the "electrostatic" and "Zn-binding" loops adjacent to the metal-binding sites are observed in the apo-enzyme relative to the fully metallated dimer. These early misfolding events expose the beta-barrels of the dimer to the external environment, allowing close interactions with adjacent molecules. Protection of the beta-edge of the protein can be partially restored by incorporating a single Zn molecule per dimer. These calculations reveal an essential step in the formation of the experimentally observed self-aggregations of metal-depleted FALS mutant SOD1. This result also has implications for the role of demetallated wild-type SOD1 in sporadic cases of ALS, for which the molecular cause still remains undiscovered.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
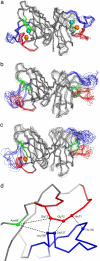
Fig. 1.
Superposition of SOD1 dimers from the starting configuration to 4 ns. (a–c) Conformational rearrangements of the electrostatic (blue) and Zn (red) loops during the MD simulations are shown by superimposing several frames for the holo- (a), apo- (b), and 1Zn/dimer (c) models. β-Strands S4 and S5 are shown in green. (d) The starting configuration of the loops is shown. The distance between the Cα atoms of residues His-71–Gly-72–Gly-73 of the Zn loop and Thr-135–Gly-127–Leu-126 of the electrostatic loop is 4.7 Å; the Asn-86 residue on β-strand 5 is 9.8 Å from Gly-73 of the Zn loop and 8.5 Å from Leu-126 of the electrostatic loop. The variation of radius of gyration over time is shown in
SI Fig. 6
.The overall separation of the loops, obtained by positional averaging over all residues, is given in
SI Fig. 7
. All figures were made by using Pymol (
).
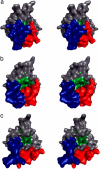
Fig. 2.
In the SOD1 monomers that lack metal, β-strands 4 and 5 become more exposed to the external environment because of movements of the loops. This is shown by surface contours of holo- (a), apo- (b), and 1Zn/dimer (c) SOD1 molecules, shown in stereo and viewed along the normal to the 2-fold dimer axis. These images were generated by using the MD trajectories at 4 ns, showing the electrostatic loop in blue, the Zn loop in red, and β-strands S4 and S5 highlighted in green. β-Strands S4 and S5 are concealed in the holo-enzyme by the closed conformation of the electrostatic and Zn-binding loops. In the apo-monomers of the apo- (b) and 1Zn/dimer (c) models, the loops adopt a more open conformation. The Asn-86 residue on β-strand S5 is clearly more exposed to the external environment. In the apo-wild-type SOD1 crystal structure (20) the Asn-86 residue interacts with the electrostatic loop of an adjacent SOD1 molecule, giving rise to an extended chain of dimer molecules in a configuration akin to the structure of amyloid-like fibrils.
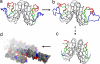
Fig. 3.
A schematic linking the MD calculations with experimental structures of apo-SOD1. (a) The experimental holo-dimer has compact Zn-binding (red) and electrostatic (blue) loops in the presence of the metals (Cu, cyan sphere; Zn, orange sphere). (b) The structure of the apo-dimer after a 4-ns MD calculation shows the disruption and increased spatial extension of the active-site loops. (c) The apo-dimer from the crystal structure (Protein Data Bank ID code 1HL4) is shown. Because the active-site loops were not visible in the electron density of this crystal structure (15) they could not be completely modeled. (d) The extended sheets of amyloid-like filaments were generated as surface contours by using the same crystal structure. The filaments consist of alternating metal-free/metal-loaded SOD1 dimers. This configuration can arise when the dimers shown in a and c are both present.
Similar articles
- Comparative structural and conformational studies on H43R and W32F mutants of copper-zinc superoxide dismutase by molecular dynamics simulation.
Muneeswaran G, Kartheeswaran S, Muthukumar K, Dharmaraj CD, Karunakaran C. Muneeswaran G, et al. Biophys Chem. 2014 Jan;185:70-8. doi: 10.1016/j.bpc.2013.11.010. Epub 2013 Dec 11. Biophys Chem. 2014. PMID: 24369116 - Unveiling the unfolding pathway of FALS associated G37R SOD1 mutant: a computational study.
Milardi D, Pappalardo M, Grasso DM, La Rosa C. Milardi D, et al. Mol Biosyst. 2010 Jun;6(6):1032-9. doi: 10.1039/b918662j. Epub 2010 Feb 23. Mol Biosyst. 2010. PMID: 20485746 - The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis.
Strange RW, Antonyuk S, Hough MA, Doucette PA, Rodriguez JA, Hart PJ, Hayward LJ, Valentine JS, Hasnain SS. Strange RW, et al. J Mol Biol. 2003 May 9;328(4):877-91. doi: 10.1016/s0022-2836(03)00355-3. J Mol Biol. 2003. PMID: 12729761 - Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis.
Rakhit R, Chakrabartty A. Rakhit R, et al. Biochim Biophys Acta. 2006 Nov-Dec;1762(11-12):1025-37. doi: 10.1016/j.bbadis.2006.05.004. Epub 2006 May 22. Biochim Biophys Acta. 2006. PMID: 16814528 Review. - Mutant SOD1 instability: implications for toxicity in amyotrophic lateral sclerosis.
Tiwari A, Hayward LJ. Tiwari A, et al. Neurodegener Dis. 2005;2(3-4):115-27. doi: 10.1159/000089616. Neurodegener Dis. 2005. PMID: 16909016 Review.
Cited by
- Copper-based pulsed dipolar ESR spectroscopy as a probe of protein conformation linked to disease states.
Merz GE, Borbat PP, Pratt AJ, Getzoff ED, Freed JH, Crane BR. Merz GE, et al. Biophys J. 2014 Oct 7;107(7):1669-74. doi: 10.1016/j.bpj.2014.07.068. Biophys J. 2014. PMID: 25296320 Free PMC article. - Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation.
Ding F, Dokholyan NV. Ding F, et al. Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19696-701. doi: 10.1073/pnas.0803266105. Epub 2008 Dec 3. Proc Natl Acad Sci U S A. 2008. PMID: 19052230 Free PMC article. - Bridging the Bridging Imidazolate in the Bimetallic Center of the Cu/Zn SOD1 and ALS.
Timucin AC, Cinaroglu SS, Sezerman OU, Timucin E. Timucin AC, et al. Front Chem. 2021 Sep 3;9:716438. doi: 10.3389/fchem.2021.716438. eCollection 2021. Front Chem. 2021. PMID: 34540798 Free PMC article. - Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS.
Roberts BR, Tainer JA, Getzoff ED, Malencik DA, Anderson SR, Bomben VC, Meyers KR, Karplus PA, Beckman JS. Roberts BR, et al. J Mol Biol. 2007 Nov 2;373(4):877-90. doi: 10.1016/j.jmb.2007.07.043. Epub 2007 Aug 2. J Mol Biol. 2007. PMID: 17888947 Free PMC article. - Structural changes to monomeric CuZn superoxide dismutase caused by the familial amyotrophic lateral sclerosis-associated mutation A4V.
Schmidlin T, Kennedy BK, Daggett V. Schmidlin T, et al. Biophys J. 2009 Sep 16;97(6):1709-18. doi: 10.1016/j.bpj.2009.06.043. Biophys J. 2009. PMID: 19751676 Free PMC article.
References
- Deng H-X, Hentati A, Tainer JA, Iqbal Z, Cayabyab A, Hung WY, Getzoff ED, Hu P, Herzfeldt B, Roos RP, et al. Science. 1993;261:1047–1051. - PubMed
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, Oregan JP, Deng HX, et al. Nature. 1993;362:59–62. - PubMed
- Cleveland DW, Rothstein JD. Nat Rev Neurosci. 2001;2:806–819. - PubMed
- Valentine JS, Doucette PA, Potter SZ. Annu Rev Biochem. 2005;74:563–593. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous