The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium - PubMed (original) (raw)
The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium
Birger Marin et al. BMC Evol Biol. 2007.
Abstract
Background: Paulinella chromatophora is a freshwater filose amoeba with photosynthetic endosymbionts (chromatophores) of cyanobacterial origin that are closely related to free-living Prochlorococcus and Synechococcus species (PS-clade). Members of the PS-clade of cyanobacteria contain a proteobacterial form 1A RubisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase) that was acquired by horizontal gene transfer (HGT) of a carboxysomal operon. In rDNA-phylogenies, the Paulinella chromatophore diverged basal to the PS-clade, raising the question whether the HGT occurred before or after the split of the chromatophore ancestor.
Results: Phylogenetic analyses of the almost complete rDNA operon with an improved taxon sampling containing most known cyanobacterial lineages recovered the Paulinella chromatophore as sister to the complete PS-clade. The sequence of the complete carboxysomal operon of Paulinella was determined. Analysis of RubisCO large subunit (rbcL) sequences revealed that Paulinella shares the proteobacterial form 1A RubisCO with the PS-clade. The gamma-proteobacterium Nitrococcus mobilis was identified as sister of the Paulinella chromatophore and the PS-clade in the RubisCO phylogeny. Gene content and order in the carboxysomal operon correlates well with the RubisCO phylogeny demonstrating that the complete carboxysomal operon was acquired by the common ancestor of the Paulinella chromatophore and the PS-clade through HGT. The carboxysomal operon shows a significantly elevated AT content in Paulinella, which in the rbcL gene is confined to third codon positions. Combined phylogenies using rbcL and the rDNA-operon resulted in a nearly fully resolved tree of the PS-clade.
Conclusion: The HGT of the carboxysomal operon predated the divergence of the chromatophore ancestor from the PS-clade. Following HGT and divergence of the chromatophore ancestor, diversification of the PS-clade into at least three subclades occurred. The gamma-proteobacterium Nitrococcus mobilis represents the closest known relative to the donor of the carboxysomal operon. The isolated position of the Paulinella chromatophore in molecular phylogenies as well as its elevated AT content suggests that the Paulinella chromatophore has already undergone typical steps in the reductive evolution of an endosymbiont.
Figures
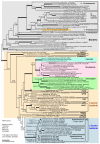
Figure 1
Phylogenetic position of the Paulinella chromatophore within the cyanobacteria inferred by complete rRNA operon sequence comparisons. The tree topology was generated by maximum likelihood (ML) analyses using the GTR+I+Γ model. The nodal support values are bootstrap values ≥ 50% obtained by ML (100 replicates), neighbor-joining (NJ; GTR+I+Γ model; 1000 repl.), maximum parsimony (MP; 1000 repl.), and Bayesian posterior probabilities (≥ 0.95). Branches in bold have maximal support (100%; 1.00) by all methods; interrupted branches were graphically reduced to 30% of their original length. Taxa in bold were newly determined for this study; strain designations and EMBL/GENBANK accession numbers are also given. Cyanobacterial clade abbreviations: GBACT (Gloeobacter), PSAN (Pseudanabaena), S/P/M (Synechocystis/Pleurocapsa/Microcystis), OSC (Oscillatoria), LEPT (Leptolyngbya), NOST (Nostoc), and PHOR (Phormidium): modified sequence groups after [17]; YELLOST (thermophilic "Synechococcus" from Yellowstone NP), THERMOSY (Thermosynechococcus), CHROO (Chroococcidiopsis), PRCHX (Prochlorothrix), SELONG (Synechococcus elongatus), and PS (Prochlorococcus/Synechococcus). Encircled numbers indicate clades/branches that were analyzed by single-gene analyses, and by NJ using the LogDet+I-model (see text and Additional File 1).
Figure 2
Synapomorphy support in the 23S rRNA for the sister-group relationship between Paulinella and free-living α-cyanobacteria. Shown is the alignment and secondary structure diagram of Helix 837 in the 23S rDNA, with two RNA base pairs highlighted that represent synapomorphies of α-cyanobacterial clades to the exclusion of Paulinella and other prokaryotes. Sequence data and evolutionary changes are plotted on a simplified phylogram (NJ-bootstrap consensus tree). Pair 868/909 shows a uniquely derived CBC (compensatory base change: U-A → C-G) of all free-living α-cyanobacteria; the neighbouring pair 869/908 changed in the common ancestor of the marine PS-subclades (marine Synechococcus and Prochlorococcus) whereas the _Cyanobium_-clade and Paulinella are plesiomorphic. Pair 869/908 shows parallel changes in a few other cyanobacteria, (e.g. in Fischerella).
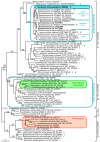
Figure 3
Evidence for HGT of RubisCO form 1A to the common ancestor of the Paulinella chromatophore and α-cyanobacteria. The ML tree was inferred from RubisCO large subunit (rbcL) form 1 amino acid sequences (470 aligned positions) of Paulinella chromatophora, cyanobacteria, plastids and proteobacteria under the RtREV+I+Γ model of amino acid substitution. Numbers at branches are ML bootstrap values ≥ 50%. Strain designations (when available) and NCBI accession numbers are indicated after the species name. Newly determined sequences are given in bold. Greek letters in grey indicate α-, β-, or γ- proteobacteria. Arrowheads highlight strains for which the gene arrangement of the carboxysomal operon is shown in Figure 5.
Figure 4
Unique synapomorphies highlighting the HGT of RubisCO from a _Nitrococcus_-like γ-proteobacterium to the common ancestor of the Paulinella chromatophore and free-living α-cyanobacteria. Selected regions of the RubisCO large subunit amino acid sequence were plotted against a simplified phylogram (NJ bootstrap consensus tree). Three synapomorphies shared by Nitrococcus and α-cyanobacteria are shown in red colour (positions 36, 59, 64); Histidine 399 and Serine 405 (blue) are unique for Paulinella and free-living α-cyanobacteria to the exclusion of all proteobacterial ancestors and β-cyanobacteria.
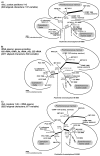
Figure 5
Architecture and evolution of operons containing form 1A RubisCO from proteobacteria and α-cyanobacteria including the Paulinella chromatophore. Gene arrangements from selected taxa (see arrowheads in Figure 3) are plotted against a simplified phylogram based on RubisCO amino acid sequences. Four major types of gene arrangements can be distinguished (for details, see text). The operon of Paulinella is member of the α-cyano-cso-type, which is derived from the ancestral cso-type present in proteobacteria, providing evidence for a HGT of the complete operon. Homologous genes share the same colour. Abbreviations: Carboxysomal shell proteins 1, 2, 3 (csoS1, 2, 3); RubisCO large and small subunit (rbcL, rbcS = cbbL, cbbS); carboxysomal peptides A, B (pepA, pepB); bacterioferritin (bfr); LysR-type transcriptional activator (cbbR); putative RubisCO activation proteins (cbbQ, cbbO); hypothetical proteins (hypo). Dotted lines indicate that no data are available.
Figure 6
Comparison of phylogenetic relationships among Paulinella and free-living α-cyanobacteria inferred by rbcL and/or rDNA nucleotide sequence data. A. Unrooted analysis of codon positions 1+2 of the rbcL gene. B. Phylogeny of the rDNA operon, using more aligned positions as in Figure 1 (4317 vs. 4126 characters). C. Phylogeny inferred from concatenated rbcL and rDNA sequences. Tree topologies resulted from ML analyses using a GTR+I+Γ model; significance values shown as in Figure 1.
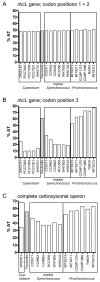
Figure 7
AT content bias in rbcL and complete carboxysomal operon sequences in Paulinella and Prochlorococcus. Whereas the AT-content in codon positions 1 + 2 in the rbcL gene is balanced across Paulinella and 17 free-living α-cyanobacteria (A), the third codon position displays a sharply elevated AT-content in Paulinella as well as Prochlorococcus strains (B). The AT-content integrated over the complete carboxysomal operon (from csoS1 to pepB) including intergenic spacer regions shows a similar bias, although less pronounced (C).
Similar articles
- Paulinella longichromatophora sp. nov., a New Marine Photosynthetic Testate Amoeba Containing a Chromatophore.
Kim S, Park MG. Kim S, et al. Protist. 2016 Feb;167(1):1-12. doi: 10.1016/j.protis.2015.11.003. Epub 2015 Nov 28. Protist. 2016. PMID: 26709891 - A single origin of the photosynthetic organelle in different Paulinella lineages.
Yoon HS, Nakayama T, Reyes-Prieto A, Andersen RA, Boo SM, Ishida K, Bhattacharya D. Yoon HS, et al. BMC Evol Biol. 2009 May 13;9:98. doi: 10.1186/1471-2148-9-98. BMC Evol Biol. 2009. PMID: 19439085 Free PMC article. - Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora.
Nowack EC, Price DC, Bhattacharya D, Singer A, Melkonian M, Grossman AR. Nowack EC, et al. Proc Natl Acad Sci U S A. 2016 Oct 25;113(43):12214-12219. doi: 10.1073/pnas.1608016113. Epub 2016 Oct 10. Proc Natl Acad Sci U S A. 2016. PMID: 27791007 Free PMC article. - Paulinella, a model for understanding plastid primary endosymbiosis.
Gabr A, Grossman AR, Bhattacharya D. Gabr A, et al. J Phycol. 2020 Aug;56(4):837-843. doi: 10.1111/jpy.13003. Epub 2020 May 5. J Phycol. 2020. PMID: 32289879 Free PMC article. Review. - Paulinella chromatophora.
Macorano L, Nowack ECM. Macorano L, et al. Curr Biol. 2021 Sep 13;31(17):R1024-R1026. doi: 10.1016/j.cub.2021.07.028. Curr Biol. 2021. PMID: 34520707 Review.
Cited by
- Endosymbiotic associations within protists.
Nowack EC, Melkonian M. Nowack EC, et al. Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):699-712. doi: 10.1098/rstb.2009.0188. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124339 Free PMC article. Review. - The CO2-concentrating mechanism of Synechococcus WH5701 is composed of native and horizontally-acquired components.
Rae BD, Förster B, Badger MR, Price GD. Rae BD, et al. Photosynth Res. 2011 Sep;109(1-3):59-72. doi: 10.1007/s11120-011-9641-5. Epub 2011 Mar 8. Photosynth Res. 2011. PMID: 21384181 - Origins of a cyanobacterial 6-phosphogluconate dehydrogenase in plastid-lacking eukaryotes.
Maruyama S, Misawa K, Iseki M, Watanabe M, Nozaki H. Maruyama S, et al. BMC Evol Biol. 2008 May 17;8:151. doi: 10.1186/1471-2148-8-151. BMC Evol Biol. 2008. PMID: 18485228 Free PMC article. - Inorganic carbon acquisition by eukaryotic algae: four current questions.
Raven JA. Raven JA. Photosynth Res. 2010 Nov;106(1-2):123-34. doi: 10.1007/s11120-010-9563-7. Epub 2010 Jun 4. Photosynth Res. 2010. PMID: 20524069 Review. - Evidence of a chimeric genome in the cyanobacterial ancestor of plastids.
Gross J, Meurer J, Bhattacharya D. Gross J, et al. BMC Evol Biol. 2008 Apr 23;8:117. doi: 10.1186/1471-2148-8-117. BMC Evol Biol. 2008. PMID: 18433492 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases