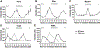Identification of the circadian transcriptome in adult mouse skeletal muscle - PubMed (original) (raw)
Identification of the circadian transcriptome in adult mouse skeletal muscle
John J McCarthy et al. Physiol Genomics. 2007.
Abstract
Circadian rhythms are approximate 24-h behavioral and physiological cycles that function to prepare an organism for daily environmental changes. The basic clock mechanism is a network of transcriptional-translational feedback loops that drive rhythmic expression of genes over a 24-h period. The objectives of this study were to identify transcripts with a circadian pattern of expression in adult skeletal muscle and to determine the effect of the Clock mutation on gene expression. Expression profiling on muscle samples collected every 4 h for 48 h was performed. Using COSOPT, we identified a total of 215 transcripts as having a circadian pattern of expression. Real-time PCR results verified the circadian expression of the core clock genes, Bmal1, Per2, and Cry2. Annotation revealed cycling genes were involved in a range of biological processes including transcription, lipid metabolism, protein degradation, ion transport, and vesicular trafficking. The tissue specificity of the skeletal muscle circadian transcriptome was highlighted by the presence of known muscle-specific genes such as Myod1, Ucp3, Atrogin1 (Fbxo32), and Myh1 (myosin heavy chain IIX). Expression profiling was also performed on muscle from the Clock mutant mouse and sarcomeric genes such as actin and titin, and many mitochondrial genes were significantly downregulated in the muscle of Clock mutant mice. Defining the circadian transcriptome in adult skeletal muscle and identifying the significant alterations in gene expression that occur in muscle of the Clock mutant mouse provide the basis for understanding the role of circadian rhythms in the daily maintenance of skeletal muscle.
Figures
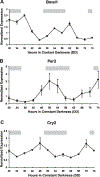
Fig. 1.
Real-time PCR verification of core clock gene expression in skeletal muscle. Real-time PCR was performed to validate the circadian expression of core clock genes identified by COSOPT from the array results. Graphical presentation of the results shows brain muscle arnt-like 1 (Bma1, A), period 2 (Per2, B), and cryptochrome (Cry2, C) transcript levels oscillate over 48 h with a circadian period indicative of a functioning circadian clock in adult skeletal muscle. Independent duplicate samples were run at each of 12 time points and normalized to GAPDH expression as described in METHODS. The light gray bars across the top of each graph represent the presumptive night with the intervening white spaces representing presumptive day.
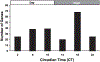
Fig. 2.
Temporal distribution of circadian genes by peak expression. Cycling genes were clustered according to their respective circadian time of peak expression. Genes were equally distributed between subjective day [circadian time 2 (CT2), CT6, and CT10: n = 103] and subjective night (CT14, CT18, and CT22: n = 112). Almost one-third (n = 68) of the cycling transcripts had peak expression at CT18, a time period of high physical activity and feeding.
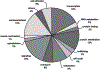
Fig. 3.
Gene ontology of skeletal muscle circadian transcriptome. A pie chart showing the distribution of circadian genes by biological process. Gene annotation was performed using GNF SymAtlas and literature searches. The “others” designation represents biological processes that occurred infrequently in the circadian transcriptome and included (frequency in parentheses) blood coagulation (2), chitin metabolism (1), DNA repair (1), meiosis (2), methyl-transferase (1), and defense response (2). The list of genes within each biological cluster is shown in Supplemental Table S1.
Fig. 4.
Altered expression of circadian genes in Clock mutant. Approximately 30% of the circadian genes in the Clock mutant showed a loss in cycling or a phase shift in peak expression. Graphs comparing wild-type vs. Clock mutant expression shows a loss of cycling (A–C) or a shift in the time of peak expression (D, E). The absence of circadian oscillation in the Clock mutant is shown for Per2 (A), Dbp (B), and Myod1 (C), three known clock-controlled genes. A phase shift in peak expression in the Clock mutant is shown for Ucp3 (D) and Pdk4 (E), both muscle-enriched circadian genes. The light gray bars across the top of each graph represent the presumptive night with the intervening white spaces representing presumptive day.
Similar articles
- CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function.
Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Andrews JL, et al. Proc Natl Acad Sci U S A. 2010 Nov 2;107(44):19090-5. doi: 10.1073/pnas.1014523107. Epub 2010 Oct 18. Proc Natl Acad Sci U S A. 2010. PMID: 20956306 Free PMC article. - Circadian rhythms, the molecular clock, and skeletal muscle.
Harfmann BD, Schroder EA, Esser KA. Harfmann BD, et al. J Biol Rhythms. 2015 Apr;30(2):84-94. doi: 10.1177/0748730414561638. Epub 2014 Dec 15. J Biol Rhythms. 2015. PMID: 25512305 Free PMC article. Review. - Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation.
Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Miller BH, et al. Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3342-7. doi: 10.1073/pnas.0611724104. Epub 2007 Feb 20. Proc Natl Acad Sci U S A. 2007. PMID: 17360649 Free PMC article. - The effect of high fat diet on daily rhythm of the core clock genes and muscle functional genes in the skeletal muscle of Chinese soft-shelled turtle (Trionyx sinensis).
Liu L, Jiang G, Peng Z, Li Y, Li J, Zou L, He Z, Wang X, Chu W. Liu L, et al. Comp Biochem Physiol B Biochem Mol Biol. 2017 Nov;213:17-27. doi: 10.1016/j.cbpb.2017.07.002. Epub 2017 Jul 17. Comp Biochem Physiol B Biochem Mol Biol. 2017. PMID: 28729066 - Circadian rhythms, the molecular clock, and skeletal muscle.
Lefta M, Wolff G, Esser KA. Lefta M, et al. Curr Top Dev Biol. 2011;96:231-71. doi: 10.1016/B978-0-12-385940-2.00009-7. Curr Top Dev Biol. 2011. PMID: 21621073 Free PMC article. Review.
Cited by
- Multitissue Circadian Proteome Atlas of WT and Per1-/-/Per2-/- Mice.
Qian L, Gu Y, Zhai Q, Xue Z, Liu Y, Li S, Zeng Y, Sun R, Zhang Q, Cai X, Ge W, Dong Z, Gao H, Zhou Y, Zhu Y, Xu Y, Guo T. Qian L, et al. Mol Cell Proteomics. 2023 Dec;22(12):100675. doi: 10.1016/j.mcpro.2023.100675. Epub 2023 Nov 7. Mol Cell Proteomics. 2023. PMID: 37940002 Free PMC article. - Chrono-Nutrition Has Potential in Preventing Age-Related Muscle Loss and Dysfunction.
Aoyama S, Nakahata Y, Shinohara K. Aoyama S, et al. Front Neurosci. 2021 Apr 16;15:659883. doi: 10.3389/fnins.2021.659883. eCollection 2021. Front Neurosci. 2021. PMID: 33935640 Free PMC article. Review. - Per1/Per2-Igf2 axis-mediated circadian regulation of myogenic differentiation.
Katoku-Kikyo N, Paatela E, Houtz DL, Lee B, Munson D, Wang X, Hussein M, Bhatia J, Lim S, Yuan C, Asakura Y, Asakura A, Kikyo N. Katoku-Kikyo N, et al. J Cell Biol. 2021 Jul 5;220(7):e202101057. doi: 10.1083/jcb.202101057. Epub 2021 May 19. J Cell Biol. 2021. PMID: 34009269 Free PMC article. - Circadian rhythms, metabolism, and insulin sensitivity: transcriptional networks in animal models.
Kitazawa M. Kitazawa M. Curr Diab Rep. 2013 Apr;13(2):223-8. doi: 10.1007/s11892-012-0354-8. Curr Diab Rep. 2013. PMID: 23266563 Review. - Deletion of Bmal1 Prevents Diet-Induced Ectopic Fat Accumulation by Controlling Oxidative Capacity in the Skeletal Muscle.
Wada T, Ichihashi Y, Suzuki E, Kosuge Y, Ishige K, Uchiyama T, Makishima M, Nakao R, Oishi K, Shimba S. Wada T, et al. Int J Mol Sci. 2018 Sep 18;19(9):2813. doi: 10.3390/ijms19092813. Int J Mol Sci. 2018. PMID: 30231537 Free PMC article.
References
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 12: 441–448, 2005. - PubMed
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–550, 2002. - PubMed
- Anderson RA, Joyce C, Davis M, Reagan JW, Clark M, Shelness GS, Rudel LL. Identification of a form of acyl-CoA:cholesterol acyltransferase specific to liver and intestine in nonhuman primates. J Biol Chem 273: 26747–26754, 1998. - PubMed
- Beatham J, Romero R, Townsend SK, Hacker T, van der Ven PF, Blanco G. Filamin C interacts with the muscular dystrophy KY protein and is abnormally distributed in mouse KY deficient muscle fibres. Hum Mol Genet 13: 2863–2874, 2004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 MH-074924/MH/NIMH NIH HHS/United States
- R21 AR050717/AR/NIAMS NIH HHS/United States
- U01 MH061915/MH/NIMH NIH HHS/United States
- R01 AR055246/AR/NIAMS NIH HHS/United States
- AR-050717/AR/NIAMS NIH HHS/United States
- P50 MH074924/MH/NIMH NIH HHS/United States
- U01 MH-61915/MH/NIMH NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources