A post-synaptic scaffold at the origin of the animal kingdom - PubMed (original) (raw)
Comparative Study
A post-synaptic scaffold at the origin of the animal kingdom
Onur Sakarya et al. PLoS One. 2007.
Abstract
Background: The evolution of complex sub-cellular structures such as the synapse requires the assembly of multiple proteins, each conferring added functionality to the integrated structure. Tracking the early evolution of synapses has not been possible without genomic information from the earliest branching animals. As the closest extant relatives to the Eumetazoa, Porifera (sponges) represent a pivotal group for understanding the evolution of nervous systems, because sponges lack neurons with clearly recognizable synapses, in contrast to eumetazoan animals.
Methodology/principal findings: We show that the genome of the demosponge Amphimedon queenslandica possesses a nearly complete set of post-synaptic protein homologs whose conserved interaction motifs suggest assembly into a complex structure. In the critical synaptic scaffold gene, dlg, residues that make hydrogen bonds and van der Waals interactions with the PDZ ligand are 100% conserved between sponge and human, as is the motif organization of the scaffolds. Expression in Amphimedon of multiple post-synaptic gene homologs in larval flask cells further supports the existence of an assembled structure. Among the few post-synaptic genes absent from Amphimedon, but present in Eumetazoa, are receptor genes including the entire ionotropic glutamate receptor family.
Conclusions/significance: Highly conserved protein interaction motifs and co-expression in sponges of multiple proteins whose homologs interact in eumetazoan synapses indicate that a complex protein scaffold was present at the origin of animals, perhaps predating nervous systems. A relatively small number of crucial innovations to this pre-existing structure may represent the founding changes that led to a post-synaptic element.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
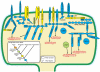
Figure 1. Origination periods of post-synaptic genes.
One possible configuration of the post-synaptic genes based on the known organization of the post-synaptic junction is illustrated. Each color represents the origination period (figure inset) of the gene family inferred from phylogenetic analyses (Figure S1). As further evidence for orthology, domain architectures of selected gene family members were compared (Figure S2). NCBI accession numbers for each gene family member are provided in Table S1. Some gene families may have been lost from the investigated genomes and originated with an earlier ancestor than shown. Question mark indicates insufficient traces to confirm this PDZ domain.
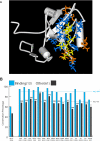
Figure 2. (A) Sponge PDZ3—CRIPT Homology Model.
The last five residues of the CRIPT protein (yellow) interact with PDZ3 residues (blue and orange) by making van der Waals contacts, hydrogen bonds, or electrostatic interactions of greater than 0.1 kcal/mol in magnitude in any of the PDZ3 homology models. (Figure S4). The subset of residues painted blue represent the core union set that interact directly with the ligand in the PDZ1 co-crystal (2I1N), the PDZ2 co-crystal (2G2L), or the PDZ3 co-crystal (1BE9) by either van der Waals contacts of 3.9 Å or shorter or by hydrogen-bond lengths of 3.5 Å or shorter. (B) Ligand-binding residues are very highly conserved within a specific type of PDZ domain. Conservation of the 13 binding residues compared to the remaining 61 more distant residues for 16 types of PDZ domains from Homo, Drosophila, Nematostella and Amphimedon. These frequencies are also calculated across all those domains at once (column 1). Comparison of the conservation of binding residues versus non-binding residues; *, p<0.05; **, p<0.01; ***, p<0.001 (Probability associated with a Student's two-sample unequal variance t-Test).
Figure 3. Expression of Post-Synaptic Orthologs in Amphimedon Larvae. DLG, GKAP, GRIP, HOMER and CRIPT.
All five genes (listed across the top) are expressed in the flask cells of Amphimedon larvae. (A–E) Sections of whole mount in situ hybridized larvae, with the posterior pole to the top. OL, outer epithelial- like layer; SL, subepithelial (middle) layer; ICM, inner cell mass. (F–J) Magnification of OL and SLs, with flask cells distributed in OL. (K–O) Scale bars: a–e, 100 µm; f–j, 50 µm; k–o, 10 µm.
Similar articles
- Co-expression of synaptic genes in the sponge Amphimedon queenslandica uncovers ancient neural submodules.
Wong E, Mölter J, Anggono V, Degnan SM, Degnan BM. Wong E, et al. Sci Rep. 2019 Oct 31;9(1):15781. doi: 10.1038/s41598-019-51282-x. Sci Rep. 2019. PMID: 31673079 Free PMC article. - The GPCR repertoire in the demosponge Amphimedon queenslandica: insights into the GPCR system at the early divergence of animals.
Krishnan A, Dnyansagar R, Almén MS, Williams MJ, Fredriksson R, Manoj N, Schiöth HB. Krishnan A, et al. BMC Evol Biol. 2014 Dec 21;14:270. doi: 10.1186/s12862-014-0270-4. BMC Evol Biol. 2014. PMID: 25528161 Free PMC article. - Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica.
Adamska M, Larroux C, Adamski M, Green K, Lovas E, Koop D, Richards GS, Zwafink C, Degnan BM. Adamska M, et al. Evol Dev. 2010 Sep-Oct;12(5):494-518. doi: 10.1111/j.1525-142X.2010.00435.x. Evol Dev. 2010. PMID: 20883218 - Sponges as models to study emergence of complex animals.
Adamska M. Adamska M. Curr Opin Genet Dev. 2016 Aug;39:21-28. doi: 10.1016/j.gde.2016.05.026. Epub 2016 Jun 16. Curr Opin Genet Dev. 2016. PMID: 27318691 Review. - Think like a sponge: The genetic signal of sensory cells in sponges.
Mah JL, Leys SP. Mah JL, et al. Dev Biol. 2017 Nov 1;431(1):93-100. doi: 10.1016/j.ydbio.2017.06.012. Epub 2017 Jun 21. Dev Biol. 2017. PMID: 28647138 Review.
Cited by
- Mutations in Nature Conferred a High Affinity Phosphatidylinositol 4,5-Bisphosphate-binding Site in Vertebrate Inwardly Rectifying Potassium Channels.
Tang QY, Larry T, Hendra K, Yamamoto E, Bell J, Cui M, Logothetis DE, Boland LM. Tang QY, et al. J Biol Chem. 2015 Jul 3;290(27):16517-29. doi: 10.1074/jbc.M115.640409. Epub 2015 May 8. J Biol Chem. 2015. PMID: 25957411 Free PMC article. - Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction.
Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Croset V, et al. PLoS Genet. 2010 Aug 19;6(8):e1001064. doi: 10.1371/journal.pgen.1001064. PLoS Genet. 2010. PMID: 20808886 Free PMC article. - Complex Homology and the Evolution of Nervous Systems.
Liebeskind BJ, Hillis DM, Zakon HH, Hofmann HA. Liebeskind BJ, et al. Trends Ecol Evol. 2016 Feb;31(2):127-135. doi: 10.1016/j.tree.2015.12.005. Epub 2015 Dec 30. Trends Ecol Evol. 2016. PMID: 26746806 Free PMC article. Review. - Where is my mind? How sponges and placozoans may have lost neural cell types.
Ryan JF, Chiodin M. Ryan JF, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Dec 19;370(1684):20150059. doi: 10.1098/rstb.2015.0059. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26554046 Free PMC article. Review. - How old is my gene?
Capra JA, Stolzer M, Durand D, Pollard KS. Capra JA, et al. Trends Genet. 2013 Nov;29(11):659-68. doi: 10.1016/j.tig.2013.07.001. Epub 2013 Aug 1. Trends Genet. 2013. PMID: 23915718 Free PMC article. Review.
References
- Gould SJ, Vrba ES. Exaptation; a missing term in the science of form Paleobiology. 1982;8(1):4–15.
- Hooper JNA, Van Soest RWM. Systema Porifera: A Guide to the classification of sponges Vols 1&2. New York: Kluwer Academic/Plenum Publishers; 2002.
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6(5):361–375. - PubMed
- Wang X, Lavrov DV. Mitochondrial Genome of the Homoscleromorph Oscarella carmela (Porifera, Demospongiae) Reveals Unexpected Complexity in the Common Ancestor of Sponges and Other Animals. Molecular biology and evolution. 2007;24(2):363–373. - PubMed
- Erpenbeck D, Voigt O, Adamski M, Adamska M, Hooper JN, et al. Mitochondrial diversity of early-branching metazoa is revealed by the complete mt genome of a haplosclerid demosponge. Molecular biology and evolution. 2007;24(1):19–22. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials