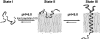A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers - PubMed (original) (raw)
A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers
Yana K Reshetnyak et al. Biophys J. 2007.
Abstract
The membrane peptide pH (low) insertion peptide (pHLIP) lives in three worlds, being soluble in aqueous solution at pH 7.4, binding to the surface of lipid bilayers, and inserting as a transbilayer helix at low pH. With low pH driving the process, pHLIP can translocate cargo molecules attached to its C-terminus via a disulfide and release them in the cytoplasm of a cell. Here we examine a key aspect of the mechanism, showing that pHLIP is monomeric in each of its three major states: soluble in water near neutral pH (state I), bound to the surface of a membrane near neutral pH (state II), and inserted across the membrane as an alpha-helix at low pH (state III). The peptide does not induce fusion or membrane leakage. The unique properties of pHLIP made it attractive for the biophysical investigation of membrane protein folding in vitro and for the development of a novel class of delivery peptides for the transport of therapeutic and diagnostic agents to acidic tissue sites associated with various pathological processes in vivo.
Figures
FIGURE 1
The three major states of pHLIP at concentration of < 30 _μ_g/mL are illustrated: soluble in water at pH > 7 (State I), bound to the surface of a lipid bilayer at the same pH and at a lipid/peptide molar ratio > 100 (State II), and inserted across the bilayer as an _α_-helix at low pH (State III).
FIGURE 2
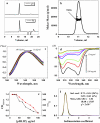
pHLIP mass in solution at pH 8.0 studied by size-exclusion chromatography coupled with on-line laser light scattering, ultraviolet, and refractive index detection. (a) The UV and RI signals of the major fraction from the column. (b) The results of SEC-LS/UV/RI analysis revealed that the major fraction from the column contained tetrameric pHLIP. Normalized fluorescence (c) and CD (d) spectra of various concentrations of pHLIP measured in 10 mM phosphate buffer, pH 8.0. The emission spectra were measured at an excitation wavelength of 295 nm. The spectral maximum shifts from 341 to 348 upon dilution. At high peptide concentration, the CD spectra have a characteristic exciton pattern (minimum at 232 nm), which disappears at low concentration where the peptide adopts a random configuration. (e) The pHLIP concentration dependence in solution of the fluorescence maximum (red point and line) and the 232:126 nm ratio of the CD signal (black point and line). (f) The distribution of sedimentation coefficients (black line) obtained by analysis of sedimentation velocity runs obtained for 30 _μ_g/mL of pHLIP in 10 mM phosphate buffer, 100 mM NaCl, pH 8.0 using the SEDFIT program. The distribution was fitted by Gaussian functions and two components were revealed (the theoretical line is red, the components are green and blue). One predominant component was found, corresponding to a peptide molecular mass of 3.84 kDa (molecular mass of the peptide is 4.1 kDa). Similar results were obtained by applying the SVEDBERG program (see text).
FIGURE 3
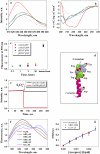
Tryptophan fluorescence (a) and CD (b) spectra of 30 _μ_g/mL pHLIP in 10 mM phosphate buffer in the absence and presence of POPC liposomes at various pHs. The fluorescence and CD spectra of pHLIP at pH 8.0 (black solid line) indicate a random configuration with tryptophan residues fully exposed to solvent. However, at low pH (pH 3.5) (black dotted line) the peptide tends to aggregate, leading to a shift of position of the fluorescence maximum and the appearance of some elements of secondary structure. Incubation of pHLIP with the POPC liposomes at pH 8.0 (blue solid line) induces the burial of tryptophan residues inside the lipid bilayer without helix formation. Decreasing the pH to 4.0 by the addition of HCl (red solid line) induces the insertion of pHLIP and the formation of helix. When POPC vesicles were added to the pHLIP at pH 3.5 there was an increase of fluorescence, a shift of the spectrum to short wavelengths, and an enhancement of helicity (red dotted line). The insertion of pHLIP across the lipid bilayer is a reversible process; an increase of pH leads to a loss of helicity and the release of the peptide from the membrane (green solid line). (c) The ability of pHLIP to induce vesicle fusion was tested on a mixture of two populations of vesicles with NDB and rhodamine fluorescently-labeled lipids. The graph presents the changes of rhodamine fluorescent signal during incubation of labeled vesicles in the presence of pHLIP at pH 8.0 and 4.0. The values were normalized to the rhodamine fluorescence (at 593 nm, excited at 450 nm) recorded after mixing of NDB-labeled liposomes with rhodamine-labeled liposomes in the absence of pHLIP. A significant increase of emission signal was observed only after several freeze-thaw cycles, which led to the fusion of vesicles. (d) An atomic representation of pHLIP taken from the crystal structure 1C3W (the C helix of bacteriorhodopsin) was generated by RasWin Molecular Graphics 2.6. The green are tryptophan residues. (e) The topology of pHLIP in a lipid bilayer was determined using the IANBD-dithionite quenching reaction. The fluorescence signal of IANBD attached to an SH group at the N-terminus (IANBD-pHLIP) or C-terminus (pHLIP-IANBD) of the peptide was monitored at 550 nm (excited at 478 nm). The starting point is a signal of IANBD-pHLIP and pHLIP-IANDB incubated with POPC followed by lowering the pH to 4.0. Then, dithionite was added. In case of IANBD conjugated to the C-terminus no change of signal is observed (black line), while the fluorescence of IANBD conjugated to the N-terminus of peptide is completely quenched (red line). The data indicate that the N-terminus of pHLIP stays outside, but that the C-terminus of pHLIP goes inside of the vesicles. (f) The pHLIP oligomeric state in a lipid bilayer was studied by FRET. The N-terminus of pHLIP was conjugated either with NDB or TAMRA. Fluorescence spectra of NDB were measured in the presence of an increasing concentration of TAMRA-pHLIP while keeping the total peptide and lipid concentration constant. (g) The comparison of calculated efficiency of energy transfer for NDB-TAMRA (blue points) and Cy3-Cy5 (red points) pairs with the theoretically predicted efficiency of energy transfer (black line) calculated from the Wolber-Hudson equation for noninteracting molecules.
Similar articles
- Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane.
Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM. Reshetnyak YK, et al. Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15340-5. doi: 10.1073/pnas.0804746105. Epub 2008 Sep 30. Proc Natl Acad Sci U S A. 2008. PMID: 18829441 Free PMC article. - Kinetics of pHLIP peptide insertion into and exit from a membrane.
Slaybaugh G, Weerakkody D, Engelman DM, Andreev OA, Reshetnyak YK. Slaybaugh G, et al. Proc Natl Acad Sci U S A. 2020 Jun 2;117(22):12095-12100. doi: 10.1073/pnas.1917857117. Epub 2020 May 14. Proc Natl Acad Sci U S A. 2020. PMID: 32409607 Free PMC article. - Comparison of lipid-dependent bilayer insertion of pHLIP and its P20G variant.
Vasquez-Montes V, Gerhart J, King KE, Thévenin D, Ladokhin AS. Vasquez-Montes V, et al. Biochim Biophys Acta Biomembr. 2018 Feb;1860(2):534-543. doi: 10.1016/j.bbamem.2017.11.006. Epub 2017 Nov 11. Biochim Biophys Acta Biomembr. 2018. PMID: 29138065 Free PMC article. - pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents.
Andreev OA, Engelman DM, Reshetnyak YK. Andreev OA, et al. Mol Membr Biol. 2010 Oct;27(7):341-52. doi: 10.3109/09687688.2010.509285. Epub 2010 Oct 13. Mol Membr Biol. 2010. PMID: 20939768 Free PMC article. Review. - GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery.
Li W, Nicol F, Szoka FC Jr. Li W, et al. Adv Drug Deliv Rev. 2004 Apr 23;56(7):967-85. doi: 10.1016/j.addr.2003.10.041. Adv Drug Deliv Rev. 2004. PMID: 15066755 Review.
Cited by
- Aspartate embedding depth affects pHLIP's insertion pKa.
Fendos J, Barrera FN, Engelman DM. Fendos J, et al. Biochemistry. 2013 Jul 9;52(27):4595-604. doi: 10.1021/bi400252k. Epub 2013 Jun 27. Biochemistry. 2013. PMID: 23721379 Free PMC article. - Residue-specific structures and membrane locations of pH-low insertion peptide by solid-state nuclear magnetic resonance.
Shu NS, Chung MS, Yao L, An M, Qiang W. Shu NS, et al. Nat Commun. 2015 Jul 21;6:7787. doi: 10.1038/ncomms8787. Nat Commun. 2015. PMID: 26195283 Free PMC article. - Ca2+ -dependent interactions between lipids and the tumor-targeting peptide pHLIP.
Vasquez-Montes V, Tyagi V, Sikorski E, Kyrychenko A, Freites JA, Thévenin D, Tobias DJ, Ladokhin AS. Vasquez-Montes V, et al. Protein Sci. 2022 Sep;31(9):e4385. doi: 10.1002/pro.4385. Protein Sci. 2022. PMID: 36040255 Free PMC article. - Tumor Microenvironment-Responsive Peptide-Based Supramolecular Drug Delivery System.
Zhang W, Yu L, Ji T, Wang C. Zhang W, et al. Front Chem. 2020 Jul 22;8:549. doi: 10.3389/fchem.2020.00549. eCollection 2020. Front Chem. 2020. PMID: 32775317 Free PMC article. Review. - pH (low) insertion peptide (pHLIP) inserts across a lipid bilayer as a helix and exits by a different path.
Andreev OA, Karabadzhak AG, Weerakkody D, Andreev GO, Engelman DM, Reshetnyak YK. Andreev OA, et al. Proc Natl Acad Sci U S A. 2010 Mar 2;107(9):4081-6. doi: 10.1073/pnas.0914330107. Epub 2010 Feb 16. Proc Natl Acad Sci U S A. 2010. PMID: 20160113 Free PMC article.
References
- Popot, J.-L., and D. M. Engelman. 2000. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69:881–922. - PubMed
- Engelman, D. M., Y. Chen, C. N. Chin, A. R. Curran, A. M. Dixon, A. D. Dupuy, A. S. Lee, U. Lehnert, E. E. Matthews, Y. K. Reshetnyak, A. Senes, and J. L. Popot. 2003. Membrane protein folding: beyond the two-stage model. FEBS Lett. 555:122–125. - PubMed
- Lehnert, U., Y. Xia, T. E. Royce, C. S. Goh, Y. Liu, A. Senes, H. Yu, Z. L. Zhang, D. M. Engelman, and M. Gerstein. 2004. Computational analysis of membrane proteins: genomic occurrence, structure prediction and helix interactions. Q. Rev. Biophys. 37:121–146. - PubMed
- Curran, A. R., and D. M. Engelman. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13:412–417. - PubMed
- Van den Berg, B., W. M. Jr. Clemons, I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature. 427:36–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM070895/GM/NIGMS NIH HHS/United States
- P20 RR016457/RR/NCRR NIH HHS/United States
- GM073857/GM/NIGMS NIH HHS/United States
- P20RR016457/RR/NCRR NIH HHS/United States
- GM070895/GM/NIGMS NIH HHS/United States
- R01 GM073857/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources