The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization - PubMed (original) (raw)
The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization
Célia Baroux et al. Plant Cell. 2007 Jun.
Abstract
The endosperm is a seed tissue unique to flowering plants. Due to its central role in nourishing and protecting the embryo, endosperm development is subject to parental conflicts and adaptive processes, which led to the evolution of parent-of-origin-dependent gene regulation. The role of higher-order chromatin organization in regulating the endosperm genome was long ignored due to technical hindrance. We developed a combination of approaches to analyze nuclear structure and chromatin organization in Arabidopsis thaliana endosperm. Endosperm nuclei showed a less condensed chromatin than other types of nuclei and a peculiar heterochromatin organization, with smaller chromocenters and additional heterochromatic foci interspersed in euchromatin. This is accompanied by a redistribution of the heterochromatin mark H3K9me1 from chromocenters toward euchromatin and interspersed heterochromatin. Thus, endosperm nuclei have a specific nuclear architecture and organization, which we interpret as a relaxed chromocenter-loop model. The analysis of endosperm with altered parental genome dosage indicated that the additional heterochromatin may be predominantly of maternal origin, suggesting differential regulation of maternal and paternal genomes, possibly linked to genome dosage regulation.
Figures
Figure 1.
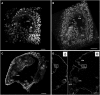
Nondenaturing Nuclear Staining in Whole-Mount Seeds. Three-dimensional reconstructions ([A] to [C]) and single plane projections (D) of propidium iodide (PI)–stained Arabidopsis seeds at stage IV ([A]; eight-nuclei endosperm stage), stage V ([B]; two-cell stage embryo delimited by the artificial contour in gray and 16-nuclei endosperm), and stage VIII ([C] and [D]; 16-cell stage embryo and ∼60-nuclei endosperm). In (C) and (D), the seed coat has been removed after cellulose and driselase treatment. (D) shows xz (left) and xy (right) plane projections of the specimen shown in (C). The frame shows a nucleus from the peripheral endosperm as used for size measurements. SC, seed coat; emb, embryo; PEN, peripheral endosperm; MCE, micropylar endosperm; CZE, chalazal endosperm; N, nucleus; mt, mitochrondria. Bars = 100 μm.
Figure 2.
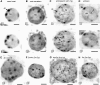
Chromosome Territories Appear More Expanded in Endosperm Than in Leaf Nuclei. Two-color fluorescence in situ hybridization (FISH) painting of chromosome 1 (top arm, red; bottom arm, green) in 2C leaf (A) and 3C nuclei from seeds at the preglobular embryo (<stage V; [B]) or later stages (stages VIII to IX; [C]), isolated by flow cytometry according to their ploidy. Chromosome territories are compact in leaf nuclei and more dispersed in endosperm nuclei. Bars = 2 μm.
Figure 3.
Endosperm Nuclei Show Specific Heterochromatic Foci Interspersed in Euchromatin That Seem Mostly Contributed by the Maternal Genome. (A) to (D) Heterochromatin-euchromatin distribution in nuclei of different origin and genetic constitution: nuclei from seed coat (A), root meristem (B), endosperm at stage IV (C), and embryo (D) from a diploid plant. (E) Sporophytic nucleus from a tetraploid plant. (F) to (H) Endosperm nuclei with different parental genome ratios as indicated (m, maternal genome; p, paternal genome) at stages IV to VI. Nuclei in (F) and (G) are derived from endosperm of reciprocal crosses between a diploid and a tetraploid line. Endosperm in (H) is derived from unfertilized seeds of the mea-2 mutant (Grossniklaus et al., 1998). PI-stained nuclei from whole-mount seeds ([A] and [C] to [H]) or seedlings ([B] and [E]) were fine-scanned along their volume under confocal microscopy. The top panels in (A) to (D) show single planes, and the bottom panels show maximum projections of three-dimensional stacks. (E) to (H) show maximum projections. cc, chromocenter; nu, nucleolus; s, chromatin strands across the nucleolus. Bars = 2 μm.
Figure 4.
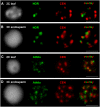
Centromeric, Athila, and 45S rDNA Repeats Are Localized at Chromocenters of Endosperm and Leaf Nuclei. FISH against centromeric repeats (CEN, red) and 45S rDNA ([A] and [B]; nucleolus-organizing regions [NOR], green) or Athila ([C] and [D]; green) in 2C leaf ([A] and [C]) and 3C endosperm nuclei from seeds with globular embryos (stages VIII to IX) ([B] and [D]), isolated by flow cytometry according to their ploidy. Left: 4′,6-diamidino-2-phenylindole (DAPI)–stained nuclei (gray). Bars = 2 μm. (A) and (B) CEN and rDNA label discrete and compact heterochromatic regions in endosperm nuclei as in leaf nuclei. (C) and (D) Athila repeats colocalize mostly but not entirely with CEN repeats in endosperm nuclei.
Figure 5.
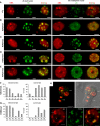
Distribution of Heterochromatin Marks in 2C Leaf and 3C Endosperm Nuclei and Specific Redistribution of H3K9me1 in Endosperm Euchromatin. (A) to (E) Indirect immunostaining of heterochromatin marks (green) and PI counterstaining (red) in 2C leaf nuclei and 3C endosperm nuclei from seeds with globular embryos (stages VIII to IX) isolated by flow cytometry according to their ploidy. H3K9me1 (B) and H3K27me2 (E) show redistribution into endosperm euchromatin compared with leaf nuclei. (F) and (G) Quantification of H3K27me2 (F) and H3K9me1 (G) immunostaining patterns in leaf and seed nuclei (stages VIII to IX) isolated by flow cytometry according to their ploidy in heterochromatin (left graph) or euchromatin (right graph). H3K27me2 redistribution toward euchromatin is found in all seed nuclei of different ploidy, while H3K9me1 redistribution is specific to endosperm nuclei (3C and 6C) (n > 100). (H) In embryos, H3K9me1 is localized to heterochromatic chromocenters. Top panels, partial projection of a quadrant-stage embryo (confocal imaging) stained with PI (red) and immunostained for H3K9me1 (green), overlaid with a picture in transmission light (right). Bottom panels, detail of an embryonic nucleus: left, DNA (PI) staining; middle, H3K9me1 immunostaining; right, overlay. emb, embryo proper; sus, suspensor; cc, chromocenters; nu, nucleolus. Bars = 2 μm.
Figure 6.
H3K9me1 Is Redistributed toward ESI Heterochromatin without Impairing Deposition of Other Euchromatin Marks. Immunostaining and PI ([A] to [F]) or DAPI (G) counterstaining of 3C seed nuclei (stages VIII to IX) isolated by flow cytometry according to their ploidy. Bars = 2 μm. (A) and (B) H3K9me1 is present as discrete foci embedded in endosperm euchromatin and colocalizes with ESI heterochromatin mostly (white arrows) but not always (black arrow). (C) to (F) The euchromatic marks H3K4me2, H3K4me3, H3K27me3, and H4K16Ac are normally distributed in endosperm nuclei. A full data set in comparison to leaf nuclei is shown in Supplemental Figure 3 online. (G) Simultaneous detection of H3K27me3 and H3K9me1.
Figure 7.
A Relaxed Chromocenter-Loop Model for Genome Organization in Endosperm Nuclei. Usually, heterochromatin in Arabidopsis nuclei is confined to chromocenters that harbor centromeric (red), pericentromeric (orange), and 45S rDNA repeats. (A) In somatic nuclei, euchromatic loops emanate from chromocenters (Fransz et al., 2002). (B) The basic loop regions normally associated with the chromocenters (black) and enriched in H3K9me1 (green) are no longer anchored at chromocenters in endosperm nuclei and may form heterochromatic, H3K9me1-rich, ESI foci. (C) Alternatively, the loops in endosperm nuclei may represent strongly decondensed chromatin with locally more condensed regions (yellow) that form ESI heterochromatin and gain H3K9me1 (green). In that case, H3K9me1 should remain associated with the chromocenters.
Similar articles
- Non-random chromosome arrangement in triploid endosperm nuclei.
Baroux C, Pecinka A, Fuchs J, Kreth G, Schubert I, Grossniklaus U. Baroux C, et al. Chromosoma. 2017 Feb;126(1):115-124. doi: 10.1007/s00412-016-0578-5. Epub 2016 Feb 19. Chromosoma. 2017. PMID: 26892012 - Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development.
Kradolfer D, Hennig L, Köhler C. Kradolfer D, et al. PLoS Genet. 2013;9(1):e1003163. doi: 10.1371/journal.pgen.1003163. Epub 2013 Jan 10. PLoS Genet. 2013. PMID: 23326241 Free PMC article. - Parent-of-origin effects on seed development in Arabidopsis thaliana.
Scott RJ, Spielman M, Bailey J, Dickinson HG. Scott RJ, et al. Development. 1998 Sep;125(17):3329-41. doi: 10.1242/dev.125.17.3329. Development. 1998. PMID: 9693137 - Heterochromatin in interphase nuclei of Arabidopsis thaliana.
Fransz P, Soppe W, Schubert I. Fransz P, et al. Chromosome Res. 2003;11(3):227-40. doi: 10.1023/a:1022835825899. Chromosome Res. 2003. PMID: 12769290 Review. - Seed evolution: parental conflicts in a multi-generational household.
Pires ND. Pires ND. Biomol Concepts. 2014 Mar;5(1):71-86. doi: 10.1515/bmc-2013-0034. Biomol Concepts. 2014. PMID: 25372743 Review.
Cited by
- Non-random chromosome arrangement in triploid endosperm nuclei.
Baroux C, Pecinka A, Fuchs J, Kreth G, Schubert I, Grossniklaus U. Baroux C, et al. Chromosoma. 2017 Feb;126(1):115-124. doi: 10.1007/s00412-016-0578-5. Epub 2016 Feb 19. Chromosoma. 2017. PMID: 26892012 - INT-Hi-C reveals distinct chromatin architecture in endosperm and leaf tissues of Arabidopsis.
Yadav VK, Santos-González J, Köhler C. Yadav VK, et al. Nucleic Acids Res. 2021 May 7;49(8):4371-4385. doi: 10.1093/nar/gkab191. Nucleic Acids Res. 2021. PMID: 33744975 Free PMC article. - An Arabidopsis AT-hook motif nuclear protein mediates somatic embryogenesis and coinciding genome duplication.
Karami O, Rahimi A, Mak P, Horstman A, Boutilier K, Compier M, van der Zaal B, Offringa R. Karami O, et al. Nat Commun. 2021 May 4;12(1):2508. doi: 10.1038/s41467-021-22815-8. Nat Commun. 2021. PMID: 33947865 Free PMC article. - Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis.
Martienssen RA. Martienssen RA. New Phytol. 2010 Apr;186(1):46-53. doi: 10.1111/j.1469-8137.2010.03193.x. New Phytol. 2010. PMID: 20409176 Free PMC article. - The SMC5/6 Complex Subunit NSE4A Is Involved in DNA Damage Repair and Seed Development.
Díaz M, Pečinková P, Nowicka A, Baroux C, Sakamoto T, Gandha PY, Jeřábková H, Matsunaga S, Grossniklaus U, Pecinka A. Díaz M, et al. Plant Cell. 2019 Jul;31(7):1579-1597. doi: 10.1105/tpc.18.00043. Epub 2019 Apr 29. Plant Cell. 2019. PMID: 31036599 Free PMC article.
References
- Adams, K.L., and Wendel, J.F. (2005). Novel patterns of gene expression in polyploid plants. Trends Genet. 21 539–543. - PubMed
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127 2493–2502. - PubMed
- Balandin, M., Royo, J., Gomez, E., Muniz, L.M., Molina, A., and Hueros, G. (2005). A protective role for the embryo surrounding region of the maize endosperm, as evidenced by the characterisation of ZmESR-6, a defensin gene specifically expressed in this region. Plant Mol. Biol. 58 269–282. - PubMed
- Baroux, C., Fransz, P., and Grossniklaus, U. (2004). Nuclear fusions contribute to polyploidization of the gigantic nuclei in the chalazal endosperm of Arabidopsis. Planta 220 38–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources