Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage - PubMed (original) (raw)
. 2007 Jun 18;177(6):969-79.
doi: 10.1083/jcb.200701042. Epub 2007 Jun 11.
Olivier J Becherel, Philip Chen, Natalie Rundle, Rick Woods, Jun Nakamura, Magtouf Gatei, Chiara Criscuolo, Alessandro Filla, Luciana Chessa, Markus Fusser, Bernd Epe, Nuri Gueven, Martin F Lavin
Affiliations
- PMID: 17562789
- PMCID: PMC2064358
- DOI: 10.1083/jcb.200701042
Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage
Amila Suraweera et al. J Cell Biol. 2007.
Abstract
A defective response to DNA damage is observed in several human autosomal recessive ataxias with oculomotor apraxia, including ataxia-telangiectasia. We report that senataxin, defective in ataxia oculomotor apraxia (AOA) type 2, is a nuclear protein involved in the DNA damage response. AOA2 cells are sensitive to H2O2, camptothecin, and mitomycin C, but not to ionizing radiation, and sensitivity was rescued with full-length SETX cDNA. AOA2 cells exhibited constitutive oxidative DNA damage and enhanced chromosomal instability in response to H2O2. Rejoining of H2O2-induced DNA double-strand breaks (DSBs) was significantly reduced in AOA2 cells compared to controls, and there was no evidence for a defect in DNA single-strand break repair. This defect in DSB repair was corrected by full-length SETX cDNA. These results provide evidence that an additional member of the autosomal recessive AOA is also characterized by a defective response to DNA damage, which may contribute to the neurodegeneration seen in this syndrome.
Figures
Figure 1.
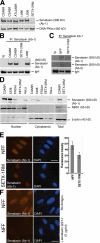
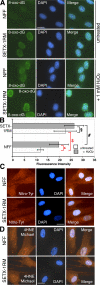
Expression and localization of senataxin. (A) Expression of senataxin in control (C2ABR and C3ABR), an unclassified AOA (ATL2ABR), AOA1 (L938, L939), and AOA2 (SETX-2RM) lymphoblastoid cells. Senataxin levels were measured in total cell extract by immunoblotting using Ab-1. DNA-PKcs is a loading control. (B) Immunoprecipitation of senataxin from C2ABR, C3ABR, L938, and SETX-2RM using Ab-1. Preimmune sera (Ig) was used as a negative control. IgH shows equal loading. (C) Immunoprecipitation of senataxin using Ab-1 from normal (NFF) and AOA2 (SETX-1RM) fibroblasts. Senataxin is detected by immunoblotting with both Ab-1 and Ab-2. IgH shows equal loading. (D) Nuclear and cytoplasmic extracts were made from C3ABR, L938, SETX-2RM, Friedrich's ataxia (FRDA1), and HeLa cells. Total cell extract from C3ABR and SETX-2RM are also shown. Senataxin levels are shown using Ab-1. NBS1 and β-actin were used to assess the purity of the fractions. (E) Cellular localization of senataxin by immunostaining of normal (NFF) and AOA2 (SETX-1RM) fibroblasts with Ab-1. DAPI shows nuclei. Fluorescence intensity from 50 individual cells for each NFF and SETX-1RM was quantitated using ImageJ software. Levels of senataxin in NFF and SETX-1RM are represented by the mean fluorescence intensity expressed in arbitrary units. SD was calculated from 50 measurements in each case. P < 0.001 (t test). (F) Confirmation of nuclear localization of senataxin using competition with antigen (senataxin GST-1), recognized by Ab-1, to inhibit binding in NFF cells. Bars, 20 μm.
Figure 2.
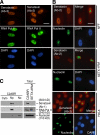
Senataxin is a nucleoplasmic protein. (A) Immunostaining for senataxin was performed in NFFs using both Ab-2 and Ab-3. RNA polymerase II identifies the nucleoplasm, and DAPI identifies the nucleus. (B) Senataxin does not localize to the nucleolus as shown by nonoverlapping staining pattern for senataxin and nucleolin, a specific marker of the nucleolus in NFFs, SETX-1RM, and HeLa. Bars, 20 μm. (C) Subcellular fractionation of control cells (C2ABR) was used to detect senataxin by immunoblotting using Ab-1. Cyto, cytoplasmic fraction; Np, nucleoplasmic fraction; No, nucleolar fraction. Detection of nucleolin and RNA polymerase II was used to determine the purity of the fractions. Total cell extract from C3ABR and SETX-2RM cells are also shown.
Figure 3.
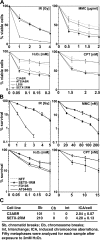
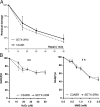
Sensitivity of AOA2 cells to DNA damaging agents. (A) Cell viability of control (C3ABR), A-T (AT25ABR), AOA1 (L939), and AOA2 (SETX-2RM) lymphoblastoid cells after treatment with the genotoxic agents (IR, MMC, H2O2, and CPT) was determined by Trypan blue staining. (B) Cell survival of normal (NFF), A-T, (AT04405) AOA1 (FD105), and AOA2 (SETX-1RM) fibroblasts after IR, MMC, CPT, and H2O2 treatments. Cell survival was determined by counting fibroblast colonies. t test analysis demonstrated a significant difference between NFF and SETX-1RM with P values of 0.008 at 0.05 mM, 0.017 at 0.1 mM, 0.014 at 0.15 mM, and 0.03 at 0.2 mM of H2O2. Error bars indicate SD. (C) Chromosome aberrations induced in response to exposure to 2 mM H2O2 for 30 min.
Figure 4.
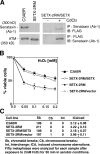
Complementation of H2O2 cell sensitivity and induced chromosome aberrations with SETX cDNA. (A) Induction of senataxin by CdCl2 in SETX-2RM cells transfected with full-length SETX cDNA. Senataxin expressed in the presence of CdCl2 was detected by immunoprecipitation with anti-senataxin followed by immunoblotting with anti-FLAG antibody or by reversing the antibody combination. (B) Complementation of cell survival of SETX-2RM lymphoblastoid with full-length SETX. Empty vector (SETX-2RM/Vector) was used as a negative control. Error bars indicate SD. (C) Complementation of H2O2-induced chromosome aberrations in SETX-2RM using full-length SETX cDNA.
Figure 5.
Evidence for oxidative damage in AOA2 fibroblasts. (A) Immunofluorescence detection of 8-oxo-dG levels in normal (NFF) and AOA2 (SETX-1RM) fibroblasts either untreated or treated with 1 mM H2O2 for 30 min in aerobic conditions. After treatment, H2O2-containing media was replaced with fresh media and cells were incubated for an additional 2 h before fixation and 8-oxo-dG detection. (B) Fluorescence intensity was quantitated on >50 individual cells for each treatment using ImageJ software. t test analysis demonstrated a significant difference in 8-oxo-dG levels in untreated NFFs and SETX-1M (*, P < 0.001) and NFFs untreated and treated with H2O2 ($, P < 0.001). No significant difference between H2O2-treated NFFs and SETX-1RM (#, P = 0.5) and SETX-1RM untreated and H2O2 treated (§, P = 0.5) was observed. Error bars indicate SD. (C) 3-nitro-tyrosine (Nitro-Tyr) staining was used to examine the status of oxidative protein damage. (D) 4-hydroxy-2-noneal (4HNE)-Michael adducts were determined to assess the levels of lipid peroxidation. DAPI shows the nuclei. Bar, 20 μm.
Figure 6.
DNA strand break repair in AOA2 cells. (A) SSB repair of control (C3ABR) and AOA2 (SETX-2RM) cells in response to H2O2-induced DNA damage measured by alkaline elution at the indicated times after treatment. SSBs were induced by treating the cells with 20 μM H2O2 for 15 min at 37°C. (B) Measure of intracellular NAD(P)H depletion in response to H2O2 and methyl methanesulfonate (MMS) treatment was also used to determine appearance of SSBs in DNA. Error bars indicate SD.
Figure 7.
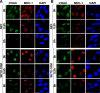
DNA damage–induced γH2AX and MDC1 foci in NFFs and AOA2 (SETX-1RM) fibroblasts. (A) H2O2-induced foci at 30 min and 8 h after treatment in control and AOA2 cells. (B) IR-induced foci at 30 min and 8 h after irradiation in control and AOA2 cells. Bars, 20 μm.
Figure 8.
Repair kinetic of DNA DSBs. (A) Quantitation of γH2AX foci in control and AOA2 cells with time after incubation with H2O2. Greater than 90% of cells had foci, and quantitation was based on >100 cells/sample in repeat experiments. t test analysis demonstrated a significant difference between NFF and SETX-1RM: *, P = 0.02; **, P = 0.003; ***, P = 0.0008. Transient transfection of GFP-SETX cDNA into AOA2 cells (green) corrected the DSB repair defect as compared with GFP-negative cells (red). (B) Quantitation of γH2AX foci in control and AOA2 cells with time after exposure of cells to 2 Gy IR. Foci were quantitated as described in A. No significant difference in the repair kinetic of γH2AX foci was observed between NNFs and SETX-1RM after IR.
Similar articles
- Aprataxin, a novel protein that protects against genotoxic stress.
Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, Gatti R, Date H, Onodera O, Taucher-Scholz G, Lavin MF. Gueven N, et al. Hum Mol Genet. 2004 May 15;13(10):1081-93. doi: 10.1093/hmg/ddh122. Epub 2004 Mar 25. Hum Mol Genet. 2004. PMID: 15044383 - A novel form of ataxia oculomotor apraxia characterized by oxidative stress and apoptosis resistance.
Gueven N, Becherel OJ, Howe O, Chen P, Haince JF, Ouellet ME, Poirier GG, Waterhouse N, Fusser M, Epe B, de Murcia JM, de Murcia G, McGowan CH, Parton R, Mothersill C, Grattan-Smith P, Lavin MF. Gueven N, et al. Cell Death Differ. 2007 Jun;14(6):1149-61. doi: 10.1038/sj.cdd.4402116. Epub 2007 Mar 9. Cell Death Differ. 2007. PMID: 17347666 - Ataxia with oculomotor apraxia type 2: clinical, biological and genotype/phenotype correlation study of a cohort of 90 patients.
Anheim M, Monga B, Fleury M, Charles P, Barbot C, Salih M, Delaunoy JP, Fritsch M, Arning L, Synofzik M, Schöls L, Sequeiros J, Goizet C, Marelli C, Le Ber I, Koht J, Gazulla J, De Bleecker J, Mukhtar M, Drouot N, Ali-Pacha L, Benhassine T, Chbicheb M, M'Zahem A, Hamri A, Chabrol B, Pouget J, Murphy R, Watanabe M, Coutinho P, Tazir M, Durr A, Brice A, Tranchant C, Koenig M. Anheim M, et al. Brain. 2009 Oct;132(Pt 10):2688-98. doi: 10.1093/brain/awp211. Epub 2009 Aug 20. Brain. 2009. PMID: 19696032 - Short-patch single-strand break repair in ataxia oculomotor apraxia-1.
Reynolds JJ, El-Khamisy SF, Caldecott KW. Reynolds JJ, et al. Biochem Soc Trans. 2009 Jun;37(Pt 3):577-81. doi: 10.1042/BST0370577. Biochem Soc Trans. 2009. PMID: 19442253 Review. - [Autosomal recessive cerebellar ataxias with oculomotor apraxia].
Le Ber I, Rivaud-Péchoux S, Brice A, Dürr A. Le Ber I, et al. Rev Neurol (Paris). 2006 Feb;162(2):177-84. doi: 10.1016/s0035-3787(06)74997-9. Rev Neurol (Paris). 2006. PMID: 16518257 Review. French.
Cited by
- XRN2 Links Transcription Termination to DNA Damage and Replication Stress.
Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, Boothman DA. Morales JC, et al. PLoS Genet. 2016 Jul 20;12(7):e1006107. doi: 10.1371/journal.pgen.1006107. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27437695 Free PMC article. - The Ighmbp2 helicase structure reveals the molecular basis for disease-causing mutations in DMSA1.
Lim SC, Bowler MW, Lai TF, Song H. Lim SC, et al. Nucleic Acids Res. 2012 Nov;40(21):11009-22. doi: 10.1093/nar/gks792. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965130 Free PMC article. - A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast.
Porrua O, Libri D. Porrua O, et al. Nat Struct Mol Biol. 2013 Jul;20(7):884-91. doi: 10.1038/nsmb.2592. Epub 2013 Jun 9. Nat Struct Mol Biol. 2013. PMID: 23748379 - Integrated genome and transcriptome analyses reveal the mechanism of genome instability in ataxia with oculomotor apraxia 2.
Kanagaraj R, Mitter R, Kantidakis T, Edwards MM, Benitez A, Chakravarty P, Fu B, Becherel O, Yang F, Lavin MF, Koren A, Stewart A, West SC. Kanagaraj R, et al. Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2114314119. doi: 10.1073/pnas.2114314119. Proc Natl Acad Sci U S A. 2022. PMID: 35042798 Free PMC article. - Defining COMMD4 as an anti-cancer therapeutic target and prognostic factor in non-small cell lung cancer.
Suraweera A, Duff A, Adams MN, Jekimovs C, Duijf PHG, Liu C, McTaggart M, Beard S, O'Byrne KJ, Richard DJ. Suraweera A, et al. Br J Cancer. 2020 Aug;123(4):591-603. doi: 10.1038/s41416-020-0899-2. Epub 2020 May 22. Br J Cancer. 2020. PMID: 32439936 Free PMC article.
References
- Ahel, I., U. Rass, S.F. El-Khamisy, S. Katyal, P.M. Clements, P.J. McKinnon, K.W. Caldecott, and S.C. West. 2006. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 443:713–716. - PubMed
- Aicardi, J., C. Barbosa, E. Andermann, F. Andermann, R. Morcos, Q. Ghanem, Y. Fukuyama, Y. Awaya, and P. Moe. 1988. Ataxia-ocular motor apraxia: a syndrome mimicking ataxia telangiectasia. Ann. Neurol. 24:497–502. - PubMed
- Bakkenist, C.J., and M.B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506. - PubMed
- Bar-Peled, M., and N.V. Raikhel. 1996. A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal. Biochem. 241:140–142. - PubMed
- Barzilai, A., G. Rotman, and Y. Shiloh. 2002. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair (Amst.). 1:3–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials