Genome-wide changes accompanying knockdown of fatty acid synthase in breast cancer - PubMed (original) (raw)
Genome-wide changes accompanying knockdown of fatty acid synthase in breast cancer
Lynn M Knowles et al. BMC Genomics. 2007.
Abstract
Background: The lipogenic enzyme fatty acid synthase (FAS) is up-regulated in a wide variety of cancers, and is considered a potential metabolic oncogene by virtue of its ability to enhance tumor cell survival. Inhibition of tumor FAS causes both cell cycle arrest and apoptosis, indicating FAS is a promising target for cancer treatment.
Results: Here, we used gene expression profiling to conduct a global study of the cellular processes affected by siRNA mediated knockdown of FAS in MDA-MB-435 mammary carcinoma cells. The study identified 169 up-regulated genes (> or = 1.5 fold) and 110 down-regulated genes (< or = 0.67 fold) in response to knockdown of FAS. These genes regulate several aspects of tumor function, including metabolism, cell survival/proliferation, DNA replication/transcription, and protein degradation. Quantitative pathway analysis using Gene Set Enrichment Analysis software further revealed that the most pronounced effect of FAS knockdown was down-regulation in pathways that regulate lipid metabolism, glycolysis, the TCA cycle and oxidative phosphorylation. These changes were coupled with up-regulation in genes involved in cell cycle arrest and death receptor mediated apoptotic pathways.
Conclusion: Together these findings reveal a wide network of pathways that are influenced in response to FAS knockdown and provide new insight into the role of this enzyme in tumor cell survival and proliferation.
Figures
Figure 1
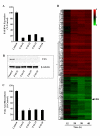
FAS knockdown generates specific and time-dependent gene expression patterns. (a-c) Target mRNA, protein and fatty acid biosynthesis knockdown by FAS siRNA duplexes. MDA-MB-435 tumor cells were exposed to 25 nM of four different siRNA duplexes targeted against FAS (FAS #1-#4) or non-silencing control siRNA for 48 h. Efficiency of FAS knockdown was determined by measuring FAS mRNA (a), FAS protein (b) and fatty acid biosynthesis (c). β-tubulin served as a loading control for FAS protein expression. Values are the mean ± SE of two replicates per treatment. (d) Expression profile represents 279 genes differentially modified by 1.5 fold in response to knockdown of FAS (shown in hours on the X-axis). Horizontal lines represent the average expression of individual genes modified by at least 3 of the FAS siRNA duplexes. Red and green indicate increased and decreased expression, respectively, relative to non-silencing control siRNA.
Figure 2
Down-regulation of metabolic pathways by targeted knockdown of FAS. The figure displays select pathways found to be down-regulated in response to FAS siRNA treatment compared to non-silencing control siRNA. Significance was determined using a nominal _p_-value < 0.05 or FDR < 0.250. The expression levels of the genes significantly modified in the pathway are coded colorimetrically: red, high expression; blue, low expression. FAS siRNA treatments for each time point are ordered as follows (a and b indicate different biological replicates): FAS #1a, FAS #1b, FAS #2a, FAS #2b, FAS #3a, FAS #3b, FAS #4a, FAS #4b. For a complete list of all pathways down-regulated by knockdown of FAS, see Additional File 6.
Figure 3
Up-regulation of cell cycle arrest and cell death pathways in response to knockdown of FAS. The figure displays select pathways found to be up-regulated in response to FAS siRNA treatment compared to non-silencing control siRNA. Significance was determined using a nominal _p_-value < 0.05 or FDR < 0.250. The expression levels of the genes significantly modified in the pathway are coded colorimetrically: red, high expression; blue, low expression. FAS siRNA treatments for each time point are ordered as follows (a and b indicate different biological replicates): FAS #1a, FAS #1b, FAS #2a, FAS #2b, FAS #3a, FAS #3b, FAS #4a, FAS #4b. For a complete list of all pathways up-regulated by knockdown of FAS, see Additional File 7.
Similar articles
- Novel signaling molecules implicated in tumor-associated fatty acid synthase-dependent breast cancer cell proliferation and survival: Role of exogenous dietary fatty acids, p53-p21WAF1/CIP1, ERK1/2 MAPK, p27KIP1, BRCA1, and NF-kappaB.
Menendez JA, Mehmi I, Atlas E, Colomer R, Lupu R. Menendez JA, et al. Int J Oncol. 2004 Mar;24(3):591-608. Int J Oncol. 2004. PMID: 14767544 - Gene expression profiling revealed survivin as a target of 3,3'-diindolylmethane-induced cell growth inhibition and apoptosis in breast cancer cells.
Rahman KW, Li Y, Wang Z, Sarkar SH, Sarkar FH. Rahman KW, et al. Cancer Res. 2006 May 1;66(9):4952-60. doi: 10.1158/0008-5472.CAN-05-3918. Cancer Res. 2006. PMID: 16651453 - Targeting fatty acid synthase in breast and endometrial cancer: An alternative to selective estrogen receptor modulators?
Lupu R, Menendez JA. Lupu R, et al. Endocrinology. 2006 Sep;147(9):4056-66. doi: 10.1210/en.2006-0486. Epub 2006 Jun 29. Endocrinology. 2006. PMID: 16809439 Review. - Progestin-induced fatty acid synthetase in breast cancer. From molecular biology to clinical applications.
Chalbos D, Joyeux C, Escot C, Galtier F, Rochefort H. Chalbos D, et al. Ann N Y Acad Sci. 1990;595:67-73. doi: 10.1111/j.1749-6632.1990.tb34283.x. Ann N Y Acad Sci. 1990. PMID: 2197971 Review. No abstract available.
Cited by
- Genome-wide linkage and association analyses implicate FASN in predisposition to Uterine Leiomyomata.
Eggert SL, Huyck KL, Somasundaram P, Kavalla R, Stewart EA, Lu AT, Painter JN, Montgomery GW, Medland SE, Nyholt DR, Treloar SA, Zondervan KT, Heath AC, Madden PA, Rose L, Buring JE, Ridker PM, Chasman DI, Martin NG, Cantor RM, Morton CC. Eggert SL, et al. Am J Hum Genet. 2012 Oct 5;91(4):621-8. doi: 10.1016/j.ajhg.2012.08.009. Am J Hum Genet. 2012. PMID: 23040493 Free PMC article. - Formation of milk lipids: a molecular perspective.
McManaman JL. McManaman JL. Clin Lipidol. 2009;4(3):391-401. doi: 10.2217/clp.09.15. Clin Lipidol. 2009. PMID: 26084294 Free PMC article. - Genome-wide analysis of YY2 versus YY1 target genes.
Chen L, Shioda T, Coser KR, Lynch MC, Yang C, Schmidt EV. Chen L, et al. Nucleic Acids Res. 2010 Jul;38(12):4011-26. doi: 10.1093/nar/gkq112. Epub 2010 Mar 9. Nucleic Acids Res. 2010. PMID: 20215434 Free PMC article. - Fatty Acid Synthase: An Emerging Target in Cancer.
Fhu CW, Ali A. Fhu CW, et al. Molecules. 2020 Aug 28;25(17):3935. doi: 10.3390/molecules25173935. Molecules. 2020. PMID: 32872164 Free PMC article. Review. - Ontology-based meta-analysis of global collections of high-throughput public data.
Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, Wall GD, Wisotzkey R, Alag S, Akhtari S, Ronaghi M. Kupershmidt I, et al. PLoS One. 2010 Sep 29;5(9):e13066. doi: 10.1371/journal.pone.0013066. PLoS One. 2010. PMID: 20927376 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous