Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation - PubMed (original) (raw)
Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation
Lothar Schermelleh et al. Nucleic Acids Res. 2007.
Abstract
Postreplicative maintenance of genomic methylation patterns was proposed to depend largely on the binding of DNA methyltransferase 1 (Dnmt1) to PCNA, a core component of the replication machinery. We investigated how the slow and discontinuous DNA methylation could be mechanistically linked with fast and processive DNA replication. Using photobleaching and quantitative live cell imaging we show that Dnmt1 binding to PCNA is highly dynamic. Activity measurements of a PCNA-binding-deficient mutant with an enzyme-trapping assay in living cells showed that this interaction accounts for a 2-fold increase in methylation efficiency. Expression of this mutant in mouse dnmt1-/- embryonic stem (ES) cells restored CpG island methylation. Thus association of Dnmt1 with the replication machinery enhances methylation efficiency, but is not strictly required for maintaining global methylation. The transient nature of this interaction accommodates the different kinetics of DNA replication and methylation while contributing to faithful propagation of epigenetic information.
Figures
Figure 1.
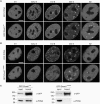
Structure of Dnmt1 and alignment of PCNA-binding domains (PBD). (A) Schematic representation of the mouse Dnmt1 consisting of an N-terminal regulatory and a C-terminal catalytic domain. The PBD, the TS domain, the Zn-binding domain (Zn), the two bromo-adjacent homology domains (BAH) and the catalytic center (PC motif) are highlighted. (B) Alignment of Dnmt1 PBDs from different species. Highly conserved residues are gray shaded. Accession numbers: Mus musculus P13864; Homo sapiens P26358; Rattus norvegicus Q9Z330; Bos taurus Q6Y856; Sus scrofa Q4TTV6; Ovis aries Q865V5; Gallus gallus Q92072; Monodelphis domestica Q8MJ28; Xenopus laevis Q6GQH0; Paracentrotus lividus Q27746; Xiphophorus maculatus Q9I8X6; Brachydanio rerio Q8QGB8. (C) Alignment of PBDs in different proteins from Mus musculus. Gray shaded amino acids are highly conserved within the PIP-Box. Accession numbers: Dnmt1 P13864; Fen1 P39749; p21 P39689; DNA Ligase1 P37913; Polymerase δ/subunit Δp66 Q9EQ28; XPG P39749; UNG:P97931 MSH6 P54276.
Figure 2.
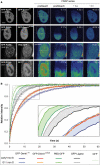
Mutation of the PBD abolishes Dnmt1 interaction with PCNA and prevents accumulation at replication sites in early and mid S-phase. (A, B) Confocal mid sections of living mouse C2C12 myoblasts expressing either GFP-Dnmt1wt (A) or GFP-Dnmt1Q162E (B). Cells were co-transfected with RFP-PCNA to identify replication foci and to distinguish S-phase stages. Scale bars: 5 µm. GFP-Dnmt1wt accumulates at replication sites throughout S phase where it co-localizes with RFP-PCNA. In G2 cells a fraction of Dnmt1 remains associated with the late replicating pericentric heterochromatin. In contrast, GFP-Dnmt1Q162E shows a fully dispersed nuclear distribution in early and mid S-phase stages, whereas in late S-phase and G2 association with centromeric heterochromatin is still observed. (C) Endogenous PCNA efficiently co-immunoprecipitates with GFP-Dnmt1wt but not with GFP-Dnmt1Q162E. Cell extracts were prepared from HEK 293T cells expressing either GFP-Dnmt1wt or GFP-Dnmt1Q162E. Precipitated proteins were detected by immunostaining with antibodies against GFP and PCNA, respectively.
Figure 3.
Transient binding of Dnmt1 at replication sites in different S-phase stages. (A) GFP-Dnmt1wt (green) and RFP-PCNA (magenta) co-expressed in mouse C2C12 cells co-localize at replication sites in early, mid and late S-phase. Fluorescence bleaching of a small region of interest reveals fast recovery of GFP-Dnmt1wt, whereas RFP-PCNA shows almost no recovery within the observation period. (B) Quantitative evaluation of the FRAP experiments shown in (A) reveal a significantly faster recovery of GFP-Dnmt1wt in early and mid S-phase compared to late S-phase. Scale bars: 5 µm.
Figure 4.
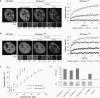
Effect of the PBD on protein mobility in S-phase cells. (A) Half nucleus FRAP series of C2C12 cells expressing GFP-Dnmt1wt and GFP-Dnmt1Q162E, PBD-GFP and GFP alone as indicated in the second column. Cell-cycle stages were identified by the subnuclear pattern of co-expressed RFP-PCNA (first column). Selected pre- and postbleach frames are shown in false color. Lower signal-to-noise ratio of FRAP series images is due to the higher imaging frame rate. Rectangles indicate the bleached ROI. The column marked with ∼_t_1/2 displays frames corresponding to the half time of fluorescence recovery. The column marked with ∼_t_∞ displays the frames corresponding to approximately the time points when fluorescence recovery reached the plateau. Bar: 5 µm. (B) Quantitative evaluation of FRAP experiments. Wild-type Dnmt1 shows a small but significant decrease in mobility in early/mid S-phase (dark blue curve) as compared to G1 phase (light-blue curve). A similar mobility shift between early/mid S and non-S-phase cells is also seen for PBD-GFP (dark green and green curve, respectively) and for GFP-Ligase (dark gray and gray curve, respectively). No such shift is observed for the PCNA-binding-deficient Dnmt1 mutant (red and orange curve, respectively). GFP alone (light-green curve) is shown as reference. For clarity only every fourth time point is displayed; data are represented as mean ± SEM. Kinetic shifts are indicated by shadings in the enlarged inset.
Figure 5.
PCNA-binding-deficient mutant Dnmt1 is catalytically active and shows moderately reduced postreplicative methylation activity in vivo. (A–C) Trapping assay for the analysis of postreplicative methylation activity in vivo. (A, B) Confocal mid sections (upper panel) and corresponding FRAP series (lower panel) of a C2C12 mouse myoblast cell co-transfected with RFP-PCNA and either GFP-Dnmt1wt (A) or GFP-Dnmt1Q162E (B) before and at selected time points after incubation with 5-aza-dC. Before drug treatment the observed cells were in early S-phase as indicated by the RFP-PCNA pattern (left). Boxes indicate bleached ROI's, which are shown magnified in the lower panels at indicated time points of the FRAP series. Quantitative FRAP analysis is shown on the right. Scale bars: 5 µm. (C) Time-dependent increase of immobile fractions of GFP-Dnmt1wt and GFP-Dnmt1Q162E. Individual cells assayed at consecutive time points are indicated by closed (GFP-Dnmt1wt) and open (GFP-Dnmt1Q162E) symbols. Character symbols represent different cells each assayed at a single time point. + symbol indicates data points obtained with C2C12 cells stably expressing GFP-Dnmt1wt (C2C12-GMT1) without co-expression of RFP-PCNA. (D) In vitro methyltransferase assay for GFP-Dnmt1wt, GFP-Dnmt1Δ1-171, GFP-Dnmt1Q162E and the catalytically inactive mutant GFP-Dnmt1C1229W. GFP fusions were expressed in HEK 293T cells, immunopurified and the amount of [3H]CH3 transferred to a hemimethylated oligonucleotide substrate was measured. To normalize for the amount of precipitated protein aliquots were analyzed by SDS-PAGE and Coomassie staining (lower panel).
Figure 6.
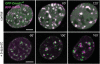
Immobilization of Dnmt1 does not prevent progression of DNA replication. S-phase C2C12 myoblasts expressing GFP-Dnmt1wt (green) and RFP-PCNA (magenta) were imaged without drug treatment (upper row) and at the indicated time points after addition of 10 µM 5-aza-dC to the medium (lower row). In the control cell the two constructs largely co-localize during transition from mid to late S-phase, whereas in the presence of 5-aza-dC progressive separation of green and red foci indicate immobilization of GFP-Dnmt1wt at postreplicative hemimethylated sites and progression of replication foci containing RFP-PCNA. Projections of confocal mid sections are shown. Scale bar: 5 µm.
Figure 7.
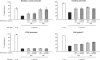
PCNA-binding-deficient mutant Dnmt1 restores methylation of single-copy sequences in dnmt1−/− ES cells. Mouse dnmt1−/− ES cells were transiently transfected with either the GFP-Dnmt1wt or GFP-Dnmt1Q162E constructs and FACS-sorted 24h and 48 h after transfection. Methylation of single-copy sequences was assayed by COBRA. Shown are methylation percentages at assayed CpG sites with respect to genomic DNA from dnmt1−/− ES cells methylated in vitro with the recombinant methyltransferase _Sss_I (100% methylation; see also Supplementary Figure 6). Mean values and standard errors of duplicate amplifications from each of two independent experiments are indicated.
Similar articles
- DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells.
Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Längst G, Leonhardt H. Spada F, et al. J Cell Biol. 2007 Feb 26;176(5):565-71. doi: 10.1083/jcb.200610062. Epub 2007 Feb 20. J Cell Biol. 2007. PMID: 17312023 Free PMC article. - The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA.
Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. Sharif J, et al. Nature. 2007 Dec 6;450(7171):908-12. doi: 10.1038/nature06397. Epub 2007 Nov 11. Nature. 2007. PMID: 17994007 - CXXC domain of human DNMT1 is essential for enzymatic activity.
Pradhan M, Estève PO, Chin HG, Samaranayke M, Kim GD, Pradhan S. Pradhan M, et al. Biochemistry. 2008 Sep 23;47(38):10000-9. doi: 10.1021/bi8011725. Epub 2008 Aug 29. Biochemistry. 2008. PMID: 18754681 - Dnmt1 structure and function.
Svedružić ŽM. Svedružić ŽM. Prog Mol Biol Transl Sci. 2011;101:221-54. doi: 10.1016/B978-0-12-387685-0.00006-8. Prog Mol Biol Transl Sci. 2011. PMID: 21507353 Review. - Non-catalytic functions of DNMT1.
Espada J. Espada J. Epigenetics. 2012 Feb;7(2):115-8. doi: 10.4161/epi.7.2.18756. Epigenetics. 2012. PMID: 22395459 Review.
Cited by
- Chromatin replication and epigenome maintenance.
Alabert C, Groth A. Alabert C, et al. Nat Rev Mol Cell Biol. 2012 Feb 23;13(3):153-67. doi: 10.1038/nrm3288. Nat Rev Mol Cell Biol. 2012. PMID: 22358331 Review. - Epigenetic regulation of retinal development and disease.
Rao RC, Hennig AK, Malik MT, Chen DF, Chen S. Rao RC, et al. J Ocul Biol Dis Infor. 2011 Sep;4(3):121-36. doi: 10.1007/s12177-012-9083-0. Epub 2012 Mar 29. J Ocul Biol Dis Infor. 2011. PMID: 23538488 Free PMC article. - Effects of diabetes-induced hyperglycemia on epigenetic modifications and DNA packaging and methylation during spermatogenesis; A narrative review.
Minas A, Camargo M, Alves MG, Bertolla RP. Minas A, et al. Iran J Basic Med Sci. 2024;27(1):3-11. doi: 10.22038/IJBMS.2023.69604.15173. Iran J Basic Med Sci. 2024. PMID: 38164482 Free PMC article. Review. - Organization of DNA replication.
Chagin VO, Stear JH, Cardoso MC. Chagin VO, et al. Cold Spring Harb Perspect Biol. 2010 Apr;2(4):a000737. doi: 10.1101/cshperspect.a000737. Cold Spring Harb Perspect Biol. 2010. PMID: 20452942 Free PMC article. Review. - Genome-wide 5-hydroxymethylcytosine (5hmC) emerges at early stage of in vitro differentiation of a putative hepatocyte progenitor.
Rodríguez-Aguilera JR, Ecsedi S, Goldsmith C, Cros MP, Domínguez-López M, Guerrero-Celis N, Pérez-Cabeza de Vaca R, Chemin I, Recillas-Targa F, Chagoya de Sánchez V, Hernández-Vargas H. Rodríguez-Aguilera JR, et al. Sci Rep. 2020 May 8;10(1):7822. doi: 10.1038/s41598-020-64700-2. Sci Rep. 2020. PMID: 32385352 Free PMC article.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. - PubMed
- Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004;61:2571–2587. - PubMed
- Lei H, Oh S, Okano M, Juttermann R, Goss K, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous