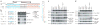Intronic microRNA precursors that bypass Drosha processing - PubMed (original) (raw)
Intronic microRNA precursors that bypass Drosha processing
J Graham Ruby et al. Nature. 2007.
Abstract
MicroRNAs (miRNAs) are approximately 22-nucleotide endogenous RNAs that often repress the expression of complementary messenger RNAs. In animals, miRNAs derive from characteristic hairpins in primary transcripts through two sequential RNase III-mediated cleavages; Drosha cleaves near the base of the stem to liberate a approximately 60-nucleotide pre-miRNA hairpin, then Dicer cleaves near the loop to generate a miRNA:miRNA* duplex. From that duplex, the mature miRNA is incorporated into the silencing complex. Here we identify an alternative pathway for miRNA biogenesis, in which certain debranched introns mimic the structural features of pre-miRNAs to enter the miRNA-processing pathway without Drosha-mediated cleavage. We call these pre-miRNAs/introns 'mirtrons', and have identified 14 mirtrons in Drosophila melanogaster and another four in Caenorhabditis elegans (including the reclassification of mir-62). Some of these have been selectively maintained during evolution with patterns of sequence conservation suggesting important regulatory functions in the animal. The abundance of introns comparable in size to pre-miRNAs appears to have created a context favourable for the emergence of mirtrons in flies and nematodes. This suggests that other lineages with many similarly sized introns probably also have mirtrons, and that the mirtron pathway could have provided an early avenue for the emergence of miRNAs before the advent of Drosha.
Figures
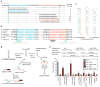
Figure 1. Introns that form pre-miRNAs
a, D. melanogaster mir-1003 with corresponding reads from high-throughput sequencing. The miRNA (red), miRNA* (blue) and splice sites (green lines) are indicated, with predicted secondary structure shown in bracket notation. b, Conservation of mir-1003 across seven Drosophila species,, coloured as in a, and also indicating consensus splice sites (green) and nucleotides differing from D. melanogaster (grey). c, Predicted secondary structures of representative debranched pre-miR-1003 orthologues, coloured as in b. d, Model for convergence of the canonical and mirtronic miRNA biogenesis pathways (see text). e, MicroRNA regulation of luciferase reporters in S2 cells. Plotted is the ratio of repression for wild-type versus mutated sites, normalized to that with the indicated non-cognate miRNA. Bar colour represents the cotransfected miRNA expression plasmid; coloured lines below indicate the cognate miRNA for the specified reporter. Error bars represent the third largest and smallest values from 12 replicates (four independent experiments, each with three transfections; *P < 0.01, **P < 0.0001, Wilcoxon rank-sum test).
Figure 2. Mirtrons are spliced as introns and diced as pre-miRNAs
a, Schematic of splice-site mutations. b, Base pairing between the indicated U1a and mir-1003 RNAs (left), and RT–PCR and northern-blot analyses of mir-1003 variants from a. The miR-1003 bands in lane 2 were attributed to endogenous miRNA. c, Northern blots analysing let-7 and mir-1003 maturation in cells treated with double-stranded RNAs (dsRNAs) corresponding to indicated genes. Shown are results from one membrane, sequentially stripped and probed for let-7 RNA, pre-miR-1003/lariat (probe 1), pre-miR-1003/miR-1003 (probe 2), and U6. Previously validated dsRNAs were used,, except for lariat debranching enzyme (CG7942, which we name ldbr), for which two unique dsRNAs were used. Knockdowns were confirmed by monitoring mRNA level and protein function (Supplementary Fig. S2). Quantification of band intensities is provided (Supplementary Table S3). *Lariat. d, Analysis of mir-1006 processing, as in c.
Figure 3. Emergence and conservation of mirtrons in species with appropriately sized introns
a, Distributions of intron (orange) and pre-miRNA (green) lengths from the indicated species. Introns and pre-miRNAs were binned by length. b, Intron and associated reads of C. elegans mir-62 (ref. 5), coloured as in Fig. 1a. Reads with untemplated nucleotides added at their 3′ terminus are shown below. c, Distributions of pre-miRNA (green) and mirtron (grey) lengths from D. melanogaster and C. elegans. d, Conservation of all 40–90-nt introns (orange) versus mirtrons (grey) from D. melanogaster (percentage identity shared with D. pseudoobscura) and C. elegans (percentage identity shared with C. briggsae).
Similar articles
- The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila.
Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. Okamura K, et al. Cell. 2007 Jul 13;130(1):89-100. doi: 10.1016/j.cell.2007.06.028. Epub 2007 Jun 28. Cell. 2007. PMID: 17599402 Free PMC article. - Mirtrons: microRNA biogenesis via splicing.
Westholm JO, Lai EC. Westholm JO, et al. Biochimie. 2011 Nov;93(11):1897-904. doi: 10.1016/j.biochi.2011.06.017. Epub 2011 Jun 21. Biochimie. 2011. PMID: 21712066 Free PMC article. Review. - The biogenesis and characterization of mammalian microRNAs of mirtron origin.
Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ. Sibley CR, et al. Nucleic Acids Res. 2012 Jan;40(1):438-48. doi: 10.1093/nar/gkr722. Epub 2011 Sep 13. Nucleic Acids Res. 2012. PMID: 21914725 Free PMC article. - Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates.
Wen J, Ladewig E, Shenker S, Mohammed J, Lai EC. Wen J, et al. PLoS Comput Biol. 2015 Sep 1;11(9):e1004441. doi: 10.1371/journal.pcbi.1004441. eCollection 2015 Sep. PLoS Comput Biol. 2015. PMID: 26325366 Free PMC article. - Versatile microRNA biogenesis in animals and their viruses.
Xie M, Steitz JA. Xie M, et al. RNA Biol. 2014;11(6):673-81. doi: 10.4161/rna.28985. Epub 2014 May 7. RNA Biol. 2014. PMID: 24823351 Free PMC article. Review.
Cited by
- A comprehensive overview on Micro RNA signature in type 2 diabetes Mellitus and its complications.
Mishra S, Bahinipati J, Sarangi R, Mohapatra SR, Das S, Mishra A. Mishra S, et al. Indian J Clin Biochem. 2023 Apr;38(2):151-158. doi: 10.1007/s12291-022-01069-1. Epub 2022 Sep 5. Indian J Clin Biochem. 2023. PMID: 36090301 Free PMC article. Review. - Swiss army knives: non-canonical functions of nuclear Drosha and Dicer.
Burger K, Gullerova M. Burger K, et al. Nat Rev Mol Cell Biol. 2015 Jul;16(7):417-30. doi: 10.1038/nrm3994. Epub 2015 May 28. Nat Rev Mol Cell Biol. 2015. PMID: 26016561 Review. - MicroRNAs in Amoebozoa: deep sequencing of the small RNA population in the social amoeba Dictyostelium discoideum reveals developmentally regulated microRNAs.
Avesson L, Reimegård J, Wagner EG, Söderbom F. Avesson L, et al. RNA. 2012 Oct;18(10):1771-82. doi: 10.1261/rna.033175.112. Epub 2012 Aug 8. RNA. 2012. PMID: 22875808 Free PMC article. - Biogenesis of mammalian microRNAs by a non-canonical processing pathway.
Havens MA, Reich AA, Duelli DM, Hastings ML. Havens MA, et al. Nucleic Acids Res. 2012 May;40(10):4626-40. doi: 10.1093/nar/gks026. Epub 2012 Jan 23. Nucleic Acids Res. 2012. PMID: 22270084 Free PMC article. - The germline of the malaria mosquito produces abundant miRNAs, endo-siRNAs, piRNAs and 29-nt small RNAs.
Castellano L, Rizzi E, Krell J, Di Cristina M, Galizi R, Mori A, Tam J, De Bellis G, Stebbing J, Crisanti A, Nolan T. Castellano L, et al. BMC Genomics. 2015 Feb 19;16(1):100. doi: 10.1186/s12864-015-1257-2. BMC Genomics. 2015. PMID: 25766668 Free PMC article.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. - PubMed
- Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous