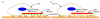The forces behind cell movement - PubMed (original) (raw)
Review
The forces behind cell movement
Revathi Ananthakrishnan et al. Int J Biol Sci. 2007.
Abstract
Cell movement is a complex phenomenon primarily driven by the actin network beneath the cell membrane, and can be divided into three general components: protrusion of the leading edge of the cell, adhesion of the leading edge and deadhesion at the cell body and rear, and cytoskeletal contraction to pull the cell forward. Each of these steps is driven by physical forces generated by unique segments of the cytoskeleton. This review examines the specific physics underlying these phases of cell movement and the origins of the forces that drive locomotion.
Conflict of interest statement
Conflict of interest: The authors have declared that no conflict of interest exists.
Figures
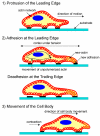
Figure 1
A schematic of the three stages of cell movement, based on ,: after determining its direction of motion, the cell extends a protusion in this direction by actin polymerization at the leading edge. It then adheres its leading edge to the surface on which it is moving and de-adheres at the cell body and rear. Finally, it pulls the whole cell body forward by contracile forces generated at the cell body and rear of the cell.
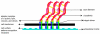
Figure 2
A schematic (based on a figure in
http://www.rpi.edu/dept/bcbp/molbiochem/MBWeb/mb2/part1/actin.htm
) showing how the cell adheres to the substrate. Cell-substrate attachments are formed when actin bundles connect to the substrate at certain sites via adhesion molecules such as vinculin, talin and integrin.
Figure 3
A schematic depicting two phenomena that can cause retrograde actin flow in vivo, based on . Retrograde flow is postulated to occur either due to release of the molecular clutch and resultant slippage, as seen in Figure a) (where the yellow and green parts of the clutch do not fit), or due to adhesion raking as shown in Figure b) (where the clutch is engaged (yellow and green parts fit) but there is raking of the cytoskeleton against the substrate (blue and white parts)).
Similar articles
- Locomotion of fish epidermal keratocytes on spatially selective adhesion patterns.
Csucs G, Quirin K, Danuser G. Csucs G, et al. Cell Motil Cytoskeleton. 2007 Nov;64(11):856-67. doi: 10.1002/cm.20230. Cell Motil Cytoskeleton. 2007. PMID: 17712861 - On the mechanisms of growth cone locomotion: modeling and computer simulation.
Li GH, Qin CD, Li MH. Li GH, et al. J Theor Biol. 1994 Aug 21;169(4):355-62. doi: 10.1006/jtbi.1994.1158. J Theor Biol. 1994. PMID: 7967628 - The comings and goings of actin: coupling protrusion and retraction in cell motility.
Small JV, Resch GP. Small JV, et al. Curr Opin Cell Biol. 2005 Oct;17(5):517-23. doi: 10.1016/j.ceb.2005.08.004. Curr Opin Cell Biol. 2005. PMID: 16099152 Review. - The cytoskeleton and brain tumour cell migration.
Maidment SL. Maidment SL. Anticancer Res. 1997 Nov-Dec;17(6B):4145-9. Anticancer Res. 1997. PMID: 9428348 Review. - Integrating adhesion, protrusion, and contraction during cell migration.
Schwartz MA, Horwitz AR. Schwartz MA, et al. Cell. 2006 Jun 30;125(7):1223-5. doi: 10.1016/j.cell.2006.06.015. Cell. 2006. PMID: 16814706
Cited by
- Cellular adhesome screen identifies critical modulators of focal adhesion dynamics, cellular traction forces and cell migration behaviour.
Fokkelman M, Balcıoğlu HE, Klip JE, Yan K, Verbeek FJ, Danen EH, van de Water B. Fokkelman M, et al. Sci Rep. 2016 Aug 17;6:31707. doi: 10.1038/srep31707. Sci Rep. 2016. PMID: 27531518 Free PMC article. - A dynamic model of chemoattractant-induced cell migration.
Yang H, Gou X, Wang Y, Fahmy TM, Leung AY, Lu J, Sun D. Yang H, et al. Biophys J. 2015 Apr 7;108(7):1645-1651. doi: 10.1016/j.bpj.2014.12.060. Biophys J. 2015. PMID: 25863056 Free PMC article. - The inhibition of lamellar hydroxyapatite and lamellar magnetic hydroxyapatite on the migration and adhesion of breast cancer cells.
Jin J, Zuo G, Xiong G, Luo H, Li Q, Ma C, Li D, Gu F, Ma Y, Wan Y. Jin J, et al. J Mater Sci Mater Med. 2014 Apr;25(4):1025-31. doi: 10.1007/s10856-013-5126-8. Epub 2013 Dec 21. J Mater Sci Mater Med. 2014. PMID: 24363068 - Scaling up single-cell mechanics to multicellular tissues - the role of the intermediate filament-desmosome network.
Broussard JA, Jaiganesh A, Zarkoob H, Conway DE, Dunn AR, Espinosa HD, Janmey PA, Green KJ. Broussard JA, et al. J Cell Sci. 2020 Mar 16;133(6):jcs228031. doi: 10.1242/jcs.228031. J Cell Sci. 2020. PMID: 32179593 Free PMC article. Review. - Analysis of the Precision, Robustness, and Speed of Elastic Resonator Interference Stress Microscopy.
Liehm P, Kronenberg NM, Gather MC. Liehm P, et al. Biophys J. 2018 May 8;114(9):2180-2193. doi: 10.1016/j.bpj.2018.03.034. Biophys J. 2018. PMID: 29742411 Free PMC article.
References
- Alberts B, Johnson A, Lewis J Molecular Biology of the Cell, 4e. Garland Science. 2002.
- Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278(5339):856–60. - PubMed
- Finer TJ, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. - PubMed
- Parekh SH, Chaudhuri O, Theriot JA et al. Loading history determines the velocity of actin-network growth. Nat Cell Biol. 2005;7(12):1219–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources