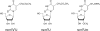A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast - PubMed (original) (raw)
A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast
Glenn R Björk et al. RNA. 2007 Aug.
Abstract
Transfer RNAs specific for Gln, Lys, and Glu from all organisms (except Mycoplasma) and organelles have a 2-thiouridine derivative (xm(5)s(2)U) as wobble nucleoside. These tRNAs read the A- and G-ending codons in the split codon boxes His/Gln, Asn/Lys, and Asp/Glu. In eukaryotic cytoplasmic tRNAs the conserved constituent (xm(5)-) in position 5 of uridine is 5-methoxycarbonylmethyl (mcm(5)). A protein (Tuc1p) from yeast resembling the bacterial protein TtcA, which is required for the synthesis of 2-thiocytidine in position 32 of the tRNA, was shown instead to be required for the synthesis of 2-thiouridine in the wobble position (position 34). Apparently, an ancient member of the TtcA family has evolved to thiolate U34 in tRNAs of organisms from the domains Eukarya and Archaea. Deletion of the TUC1 gene together with a deletion of the ELP3 gene, which results in the lack of the mcm(5) side chain, removes all modifications from the wobble uridine derivatives of the cytoplasmic tRNAs specific for Gln, Lys, and Glu, and is lethal to the cell. Since excess of the unmodified form of these three tRNAs rescued the double mutant elp3 tuc1, the primary function of mcm(5)s(2)U34 seems to be to improve the efficiency to read the cognate codons rather than to prevent mis-sense errors. Surprisingly, overexpression of the mcm(5)s(2)U-lacking tRNA(Lys) alone was sufficient to restore viability of the double mutant.
Figures
FIGURE 1.
Modified wobble uridines present in yeast tRNA reading twofold degenerate codon boxes (Fig. 2). 5-methylcarboxymethyl-2-thiouridine (mcm5s2U), 5-methylcarboxymethyl-uridine (mcm5U), and 5-carbamoylmethyl-2′-_O_-methyluridine (ncm5Um).
FIGURE 2.
Modified nucleosides in the wobble position and the coding capacities of the corresponding tRNAs. Letters outside the box, to the left, at top, and to the right indicate the first, second, and third position of the codon, respectively. Circles connected by a line, or a single circle, represent one tRNA species. A filled circle indicates the capability of that tRNA to base pair with a particular codon, either by Watson–Crick or by wobble according to the current wobble rules (Yokoyama and Nishimura 1995). An open circle suggests a restricted base pairing. To the right of each tRNA, the modification at positions 34 (the wobble position) is shown. Data compiled from Johansson and Byström (2005). Thus, tRNA specific for Lys, having mcm5s2U as wobble nucleoside, pairs well with AAA (•) but less well with AAG (○). (A) Presence of mcm5s2U in tRNAGln and mcm5U in tRNAGly as wobble nucleosides has been determined (Lu et al. 2005) (M.J.O. Johansson, pers. comm.) (B) tRNAs specific for Ser, Thr, and Ala have ncm5U, which synthesis is dependent on ELP3 (M.J.O. Johansson, pers. comm.).
FIGURE 3.
Sequence alignment of four TtcA and four Tuc1p homologs and six MnmA/Mtu1p proteins. Letters shaded black, dark-blue, and light-blue indicate that the indicated amino acid is found in this position in at least 100%, 55%, and 25% of the sequences in the alignment, respectively. Letters shaded red indicate conserved cysteine residues. Cys-X1-X2 -Cys motifs in the central domain of Groups 1 and 2 and in the flanking regions in Group 2 are, consequently, seen as two red stripes close to each other. Conserved regions thought to be important for binding and catalytic function are boxed in red. Amino acids identified by Numata et al. (2006) important for binding of the tRNA substrate (black dots) and for catalytic function (open circles), as well as the two sulphur coordinating cysteines (C102 and C199 in E. coli MnmA, red dots) are indicated. At bottom is shown a schematic comparison of Group 1 (TtcA) and Group 2 (Ygl211p, renamed to Tuc1p, see text) as suggested by Jäger et al. (2004). The central part of MnmA, which is required for the synthesis of s2U34 in bacteria, is distinctly different from Groups 1 and 2 and is similar to the yeast protein Mtu1p, which is responsible for s2U34 formation in mitochondria (Umeda et al. 2005). The central domains of both Group 1 and Group 2 consist of the PP-loop (SGGxDS) motif, a Cys-X1-X2-Cys motif, a GH motif, and a PL motif. In addition to the conserved central domain, Group 2 proteins have two Cys-X1-X2-Cys motifs on each side of the central domain. Note that MnmA has the PP-loop motif and GH motif, but otherwise is not similar to any of the TtcA family proteins, but instead has conserved regions absent in Groups 1/2 thiolases.
FIGURE 4.
Increased expression of tRNAs normally having mcm5s2U suppresses lethality induced by lack of this modification of wobble uridines. (A) Strains containing the elp3∷KanMX4 tuc1∷TRP1 mutations and carrying the URA3 plasmid pRS316-ELP3 and the LEU2 plasmids pRS425, pRS315-ELP3, pRS-425-tE(UUC), pRS425-tQ(UUG), pRS425-tK(UUU), pRS425-tK(UUU)-tQ(UUG), pRS425-tK(UUU)-tE(UUC), or pRS425-_tE(UUC)-tK(UUU)_-tQ(UUG) (tE, tK and tQ indicate the genes for tRNAGlu mcm5s2UUC, tRNALys mcm5s2UUU, and tRNAGln mcm5s2UUG, respectively), were grown in synthetic complete medium lacking leucine (SC-leu). Strains were diluted to 3 × 107 cells/mL, 10-fold serially diluted, and 5 μL was spotted onto either SC-leu plates or SC-leu plates containing 5-fluoroorotic acid (5-FOA). These plates were incubated at 30°C for 2 and 3 d, respectively. Cells containing a URA3 plasmid are unable to grow on 5-FOA-containing medium, with the result that these strains only contain the indicated LEU2 vectors when plated on 5-FOA plates (Boeke et al. 1984). (B) Growth of wild-type (W303-1B) and of the double mutant elp3∷KanMX4 tuc1∷TRP1 containing plasmids harboring genes encoding various tRNAs on SC-plates lacking leucine. Cells were picked from SC-Leu+5-FOA plates (Fig. 4A) and restreaked on SC plates lacking Leu. Plates were incubated at 30°C or at 38°C for 2.5 d. Plasmid pRS315 and pRS425 are denoted lc (low copy) and hc (high copy), respectively. (C) Growth of wild-type (W303–1B), tuc1∷TRP1 (YMP006), elp3∷KanMX4 (UMY2843), or the double mutant tuc1∷TRP1, elp3∷KanMX4/ pRS425-tEKQ (UMY3400) on rich medium (YEPD). Plasmid pRS425-tEKQ is denoted hc-tEKQ and its presence results in overexpression of tRNAGlu mcm5s2UUC, tRNALys mcm5s2UUU, and tRNAGln mcm5s2UUG. Growth at 30°C was scored after 1 or 2 d.
FIGURE 5.
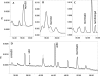
HPLC analysis of modified nucleosides of tRNA from W303-1B (wild-type; thick line) and from strain UMY3400 (_elp3∷KanMX4 tuc1∷TRP/pRS425-tE_[_UUC_]-_tK_[_UUU_]-_tQ_[_UUG_]) (tE, tK and tQ indicate the genes for tRNAGlu mcm5s2UUC, tRNALys mcm5s2UUU, and tRNAGln mcm5s2UUG; thin line). Transfer RNA was degraded to nucleosides and the distribution of nucleosides was analyzed by HPLC. (A–C) The 13–19 min region, 37–41 min region, and 49.5–54 min region, respectively, of the HPLC chromatogram monitored at 254 nm. (D) The region 30–58 min was monitored at 314 nm. The expected position of s2U is indicated at bottom, and this compound was observed only in tRNA from UMY2843 (elp3∷KanMX4). The identities of cm5s2U and ncm5s2UmpA are tentative, since no markers for these compounds were available.
FIGURE 6.
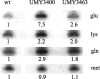
Levels of tRNAGlu mcm5s2UUC, tRNAGln mcm5s2UUG, and tRNALys mcm5s2UUU in strains W303-1B (wt), UMY3400 (_elp3∷KanMX4 tuc1∷TRP1/_pRS425-tE(UUC)-tK(UUU)-tQ(UUG)), and UMY3463 (_ELP3 TUC1/_pRS425-tE(UUC)-tK(UUU)-tQ(UUG)). The tRNA levels were monitored by Northern hybridization using radioactive oligonucleotides complementary to nucleotides 57–72 of tRNAGln, 57–73 of tRNALys, 52–72 of tRNAGlu, and 30–49 of tRNAMet. Use of the last oligonucleotide, which monitors the levels of a tRNA not having mcm5s2U, served as controls for how much material was loaded on the gel. The numbers below each lane represent the levels relative to the level of the corresponding tRNAs in strain W303-1B (wild type).
Similar articles
- The primary target of the killer toxin from Pichia acaciae is tRNA(Gln).
Klassen R, Paluszynski JP, Wemhoff S, Pfeiffer A, Fricke J, Meinhardt F. Klassen R, et al. Mol Microbiol. 2008 Aug;69(3):681-97. doi: 10.1111/j.1365-2958.2008.06319.x. Epub 2008 Jun 28. Mol Microbiol. 2008. PMID: 18532979 - MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli.
Kambampati R, Lauhon CT. Kambampati R, et al. Biochemistry. 2003 Feb 4;42(4):1109-17. doi: 10.1021/bi026536+. Biochemistry. 2003. PMID: 12549933 - Structural and mechanistic basis for enhanced translational efficiency by 2-thiouridine at the tRNA anticodon wobble position.
Rodriguez-Hernandez A, Spears JL, Gaston KW, Limbach PA, Gamper H, Hou YM, Kaiser R, Agris PF, Perona JJ. Rodriguez-Hernandez A, et al. J Mol Biol. 2013 Oct 23;425(20):3888-906. doi: 10.1016/j.jmb.2013.05.018. Epub 2013 May 28. J Mol Biol. 2013. PMID: 23727144 Free PMC article. - Aminoacylation of transfer RNAs with 2-thiouridine derivatives in the wobble position of the anticodon.
Rogers KC, Crescenzo AT, Söll D. Rogers KC, et al. Biochimie. 1995;77(1-2):66-74. doi: 10.1016/0300-9084(96)88106-5. Biochimie. 1995. PMID: 7541255 Review. - Sulfur Modifications of the Wobble U34 in tRNAs and their Intracellular Localization in Eukaryotic Cells.
Nakai Y, Nakai M, Yano T. Nakai Y, et al. Biomolecules. 2017 Feb 18;7(1):17. doi: 10.3390/biom7010017. Biomolecules. 2017. PMID: 28218716 Free PMC article. Review.
Cited by
- Mtu1-Mediated Thiouridine Formation of Mitochondrial tRNAs Is Required for Mitochondrial Translation and Is Involved in Reversible Infantile Liver Injury.
Wu Y, Wei FY, Kawarada L, Suzuki T, Araki K, Komohara Y, Fujimura A, Kaitsuka T, Takeya M, Oike Y, Suzuki T, Tomizawa K. Wu Y, et al. PLoS Genet. 2016 Sep 30;12(9):e1006355. doi: 10.1371/journal.pgen.1006355. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27689697 Free PMC article. - The cytosolic thiouridylase CTU2 of Arabidopsis thaliana is essential for posttranscriptional thiolation of tRNAs and influences root development.
Philipp M, John F, Ringli C. Philipp M, et al. BMC Plant Biol. 2014 Apr 28;14:109. doi: 10.1186/1471-2229-14-109. BMC Plant Biol. 2014. PMID: 24774365 Free PMC article. - Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation.
Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. Deng W, et al. PLoS Genet. 2015 Dec 15;11(12):e1005706. doi: 10.1371/journal.pgen.1005706. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26670883 Free PMC article. - Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae.
Hopper AK. Hopper AK. Genetics. 2013 May;194(1):43-67. doi: 10.1534/genetics.112.147470. Genetics. 2013. PMID: 23633143 Free PMC article. Review. - Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity.
Nedialkova DD, Leidel SA. Nedialkova DD, et al. Cell. 2015 Jun 18;161(7):1606-18. doi: 10.1016/j.cell.2015.05.022. Epub 2015 Jun 4. Cell. 2015. PMID: 26052047 Free PMC article.
References
- Agris, P.F., Vendeix, F.A., Graham, W.D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2006;366:1–13. - PubMed
- Avital, S., Elson, D. A convenient procedure for preparing transfer ribonucleic acid from Escherichia coli . Biochim. Biophys. Acta. 1969;179:297–307. - PubMed
- Björk, G.R. Modified nucleosides in positions 34 and 37 of tRNAs and their predicted coding capacities. In: Grosjean H., Benne B., editors. Modification and editing of RNA. American Society for Microbiology; Washington, DC: 1998. pp. 577–581.
- Björk, G.R., Hagervall, T.G. 25 July 2005, posting date. Chapter 4.6.2. Transfer RNA modification. In Escherichia coli and Salmonella. In: Böck A., et al., editors. Cellular and molecular biology. ASM Press; Washington, DC: 2005.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases