Indexing strategies for rapid searches of short words in genome sequences - PubMed (original) (raw)
Indexing strategies for rapid searches of short words in genome sequences
Christian Iseli et al. PLoS One. 2007.
Abstract
Searching for matches between large collections of short (14-30 nucleotides) words and sequence databases comprising full genomes or transcriptomes is a common task in biological sequence analysis. We investigated the performance of simple indexing strategies for handling such tasks and developed two programs, fetchGWI and tagger, that index either the database or the query set. Either strategy outperforms megablast for searches with more than 10,000 probes. FetchGWI is shown to be a versatile tool for rapidly searching multiple genomes, whose performance is limited in most cases by the speed of access to the filesystem. We have made publicly available a Web interface for searching the human, mouse, and several other genomes and transcriptomes with oligonucleotide queries.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
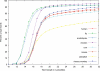
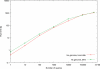
Figure 1. Word duplication in several genomes.
Analysis of the percentage of unique sequence tags in several genomes as a function of tag length.
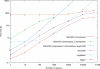
Figure 2. Runtime comparisons on the human genome.
Runtime comparisons between fetchGWI using plain and compressed index files, tagger, and megablast. Each point is computed from the average of three runs on the human genome with different input data, except the last run done on the whole dataset. Only perfect matches are sought, except for the 2 experiments explicitly noted were 2 mismatches were allowed.
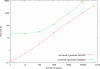
Figure 3. Runtime comparisons on multiple genomes.
Runtime comparisons between fetchGWI (using a compressed index file) and megablast. Each point is computed from the average of three runs on the combined genome of 9 species (human, mouse, honey bee, cattle, dog, drosophila, zebrafish, chimp, and rat) with different input data, except the last run done on the whole dataset. Only perfect matches were sought.
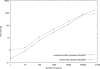
Figure 4. Runtime comparisons on combined index files.
Runtime comparisons of fetchGWI when using either multiple index files, or a single, combined, index file. Each point is computed from the average of three runs on the human and mouse genomes with different input data, except the last run done on the whole dataset Only perfect matches were sought.
Figure 5. Runtime comparisons on different filesystems.
Runtime comparisons of fetchGWI when the index file is stored either on a filesystem on local disks, or on a SFS cluster filesystem. Each point is computed from the average of three runs with different input data, except the last run done on the whole dataset. Only perfect matches were sought.
Similar articles
- Database indexing for production MegaBLAST searches.
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Morgulis A, et al. Bioinformatics. 2008 Aug 15;24(16):1757-64. doi: 10.1093/bioinformatics/btn322. Epub 2008 Jun 21. Bioinformatics. 2008. PMID: 18567917 Free PMC article. - Indexing and searching petabase-scale nucleotide resources.
Shiryev SA, Agarwala R. Shiryev SA, et al. Nat Methods. 2024 Jun;21(6):994-1002. doi: 10.1038/s41592-024-02280-z. Epub 2024 May 16. Nat Methods. 2024. PMID: 38755321 Free PMC article. - MICA: desktop software for comprehensive searching of DNA databases.
Stokes WA, Glick BS. Stokes WA, et al. BMC Bioinformatics. 2006 Oct 3;7:427. doi: 10.1186/1471-2105-7-427. BMC Bioinformatics. 2006. PMID: 17018144 Free PMC article. - Databases and software for the comparison of prokaryotic genomes.
Field D, Feil EJ, Wilson GA. Field D, et al. Microbiology (Reading). 2005 Jul;151(Pt 7):2125-2132. doi: 10.1099/mic.0.28006-0. Microbiology (Reading). 2005. PMID: 16000703 Review. - [Transcriptomes for serial analysis of gene expression].
Marti J, Piquemal D, Manchon L, Commes T. Marti J, et al. J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French.
Cited by
- Detection of genomic variation by selection of a 9 mb DNA region and high throughput sequencing.
Nikolaev SI, Iseli C, Sharp AJ, Robyr D, Rougemont J, Gehrig C, Farinelli L, Antonarakis SE. Nikolaev SI, et al. PLoS One. 2009 Aug 17;4(8):e6659. doi: 10.1371/journal.pone.0006659. PLoS One. 2009. PMID: 19684856 Free PMC article. - One-step generation of phenotype-expressing triple-knockout mice with heritable mutated alleles by the CRISPR/Cas9 system.
Fujii W, Onuma A, Sugiura K, Naito K. Fujii W, et al. J Reprod Dev. 2014;60(4):324-7. doi: 10.1262/jrd.2013-139. Epub 2014 May 4. J Reprod Dev. 2014. PMID: 25110137 Free PMC article. - Effect of C-type lectin 16 on dengue virus infection in Aedes aegypti salivary glands.
Chang YC, Liu WL, Fang PH, Li JC, Liu KL, Huang JL, Chen HW, Kao CF, Chen CH. Chang YC, et al. PNAS Nexus. 2024 May 16;3(5):pgae188. doi: 10.1093/pnasnexus/pgae188. eCollection 2024 May. PNAS Nexus. 2024. PMID: 38813522 Free PMC article. - GapmeR cellular internalization by macropinocytosis induces sequence-specific gene silencing in human primary T-cells.
Fazil MH, Ong ST, Chalasani ML, Low JH, Kizhakeyil A, Mamidi A, Lim CF, Wright GD, Lakshminarayanan R, Kelleher D, Verma NK. Fazil MH, et al. Sci Rep. 2016 Nov 24;6:37721. doi: 10.1038/srep37721. Sci Rep. 2016. PMID: 27883055 Free PMC article. - Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations.
Khan SA, Reichelt M, Heckel DG. Khan SA, et al. Sci Rep. 2017 Jan 5;7:40025. doi: 10.1038/srep40025. Sci Rep. 2017. PMID: 28053351 Free PMC article.
References
- Lal A, Sui IM, Riggins G. Serial analysis of gene expression: Probing transcriptomes for molecular targets. Current Opinion in Molecular Therapeutics. 1999;1:720–726. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources