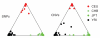The population genetics of structural variation - PubMed (original) (raw)
Review
. 2007 Jul;39(7 Suppl):S30-6.
doi: 10.1038/ng2042.
Affiliations
- PMID: 17597779
- PMCID: PMC2716079
- DOI: 10.1038/ng2042
Review
The population genetics of structural variation
Donald F Conrad et al. Nat Genet. 2007 Jul.
Abstract
Population genetics is central to our understanding of human variation, and by linking medical and evolutionary themes, it enables us to understand the origins and impacts of our genomic differences. Despite current limitations in our knowledge of the locations, sizes and mutational origins of structural variants, our characterization of their population genetics is developing apace, bringing new insights into recent human adaptation, genome biology and disease. We summarize recent dramatic advances, describe the diverse mutational origins of chromosomal rearrangements and argue that their complexity necessitates a re-evaluation of existing population genetic methods.
Figures
Figure 1
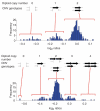
Diploid copy numbers, corresponding CNV genotypes and the underlying quantitative data from an array CGH experiment. Top, biallelic CNV; bottom, multiallelic CNV (data from ref. 11). Note that there is not a 1:1 mapping of diploid copy numbers to CNV genotypes for the multiallelic CNV.
Figure 2
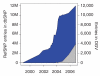
Cumulative number of RefSNP entries in dbSNP and cumulative number of variant loci in the Database of Genomic Variants, plotted as a function of time. Axes have been scaled differently to enhance visualization. RefSNP entries: left axis, blue (M = million). DGV entries: right axis, gray.
Figure 3
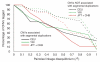
The maximal pairwise LD with a nearby SNP is lower around CNVs that are associated with segmental duplications than around CNVs in single-copy sequences.
Figure 4
Estimates of fine-scale recombination rate across the 8p23 inversion. Haplotypes from Supplementary Figure 2 were used to generate estimates of population genetic recombination rate using the method from ref. . Phylogenetic analysis of the SNP haplotypes uncovered two primary clades, and clade membership was used as a proxy for inversion status, with the minor allele assumed to be the inverted allele, arbitrarily. (a) Estimate of the population-scaled recombination rate (ρ = 4_N_e_r_) using 20 minor and 20 major (common) alleles. (b) Estimate of 4_N_e_r_ using 40 haplotypes of the minor allele. (c) Estimate of 4_N_e_r_ using 40 haplotypes of the major allele. As recombination is restricted in inversion heterozygotes, mutations that are private to either inversion background will be in extreme LD when considered at the population level, leading to low estimates of 4_N_e_r_ in a. The analysis for a was run several times with different sets of minor and major haplotypes, and at most one ‘hotspot’ was detected visually.
Figure 5
Plots of population structure for 67 CNVs and 67 unlinked SNPs in 210 unrelated HapMap individuals, assuming three ancestral populations. The slightly improved clustering quality from SNP genotypes most likely relates to the lower frequency of missing genotypes in the SNP data.
Similar articles
- Population genetics. Seeking the signs of selection.
Olson S. Olson S. Science. 2002 Nov 15;298(5597):1324-5. doi: 10.1126/science.298.5597.1324. Science. 2002. PMID: 12434033 No abstract available. - Structural variants deconstruct the genome.
[No authors listed] [No authors listed] Nat Genet. 2006 Sep;38(9):959. doi: 10.1038/ng0906-959. Nat Genet. 2006. PMID: 16940994 - Population genetic nature of copy number variation.
Sjödin P, Jakobsson M. Sjödin P, et al. Methods Mol Biol. 2012;838:209-23. doi: 10.1007/978-1-61779-507-7_10. Methods Mol Biol. 2012. PMID: 22228014 Review. - Identification of copy number variants defining genomic differences among major human groups.
Armengol L, Villatoro S, González JR, Pantano L, García-Aragonés M, Rabionet R, Cáceres M, Estivill X. Armengol L, et al. PLoS One. 2009 Sep 30;4(9):e7230. doi: 10.1371/journal.pone.0007230. PLoS One. 2009. PMID: 19789632 Free PMC article. - Structure of linkage disequilibrium in humans: genome factors and population stratification.
Bertranpetit J, Calafell F, Comas D, González-Neira A, Navarro A. Bertranpetit J, et al. Cold Spring Harb Symp Quant Biol. 2003;68:79-88. doi: 10.1101/sqb.2003.68.79. Cold Spring Harb Symp Quant Biol. 2003. PMID: 15338606 Review. No abstract available.
Cited by
- Deleterious Mutations and the Rare Allele Burden on Rice Gene Expression.
Lye Z, Choi JY, Purugganan MD. Lye Z, et al. Mol Biol Evol. 2022 Sep 1;39(9):msac193. doi: 10.1093/molbev/msac193. Mol Biol Evol. 2022. PMID: 36073358 Free PMC article. - Whole genome distribution and ethnic differentiation of copy number variation in Caucasian and Asian populations.
Li J, Yang T, Wang L, Yan H, Zhang Y, Guo Y, Pan F, Zhang Z, Peng Y, Zhou Q, He L, Zhu X, Deng H, Levy S, Papasian CJ, Drees BM, Hamilton JJ, Recker RR, Cheng J, Deng HW. Li J, et al. PLoS One. 2009 Nov 23;4(11):e7958. doi: 10.1371/journal.pone.0007958. PLoS One. 2009. PMID: 19956714 Free PMC article. - Population-genetic nature of copy number variations in the human genome.
Kato M, Kawaguchi T, Ishikawa S, Umeda T, Nakamichi R, Shapero MH, Jones KW, Nakamura Y, Aburatani H, Tsunoda T. Kato M, et al. Hum Mol Genet. 2010 Mar 1;19(5):761-73. doi: 10.1093/hmg/ddp541. Epub 2009 Dec 5. Hum Mol Genet. 2010. PMID: 19966329 Free PMC article. - Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis.
Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Ionita-Laza I, et al. Genomics. 2009 Jan;93(1):22-6. doi: 10.1016/j.ygeno.2008.08.012. Epub 2008 Oct 19. Genomics. 2009. PMID: 18822366 Free PMC article. Review. - MOCSphaser: a haplotype inference tool from a mixture of copy number variation and single nucleotide polymorphism data.
Kato M, Nakamura Y, Tsunoda T. Kato M, et al. Bioinformatics. 2008 Jul 15;24(14):1645-6. doi: 10.1093/bioinformatics/btn242. Epub 2008 May 20. Bioinformatics. 2008. PMID: 18492685 Free PMC article.
References
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. - PubMed
- Jobling MA, Hurles ME, Tyler-Smith C. Human Evolutionary Genetics: Origins, Peoples and Disease. Garland Science; New York: 2004.
- Flint J, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321:744–750. - PubMed
- Conrad DF, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 2006;38:1251–1260. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources