Genome-wide maps of chromatin state in pluripotent and lineage-committed cells - PubMed (original) (raw)
. 2007 Aug 2;448(7153):553-60.
doi: 10.1038/nature06008. Epub 2007 Jul 1.
Manching Ku, David B Jaffe, Biju Issac, Erez Lieberman, Georgia Giannoukos, Pablo Alvarez, William Brockman, Tae-Kyung Kim, Richard P Koche, William Lee, Eric Mendenhall, Aisling O'Donovan, Aviva Presser, Carsten Russ, Xiaohui Xie, Alexander Meissner, Marius Wernig, Rudolf Jaenisch, Chad Nusbaum, Eric S Lander, Bradley E Bernstein
Affiliations
- PMID: 17603471
- PMCID: PMC2921165
- DOI: 10.1038/nature06008
Genome-wide maps of chromatin state in pluripotent and lineage-committed cells
Tarjei S Mikkelsen et al. Nature. 2007.
Abstract
We report the application of single-molecule-based sequencing technology for high-throughput profiling of histone modifications in mammalian cells. By obtaining over four billion bases of sequence from chromatin immunoprecipitated DNA, we generated genome-wide chromatin-state maps of mouse embryonic stem cells, neural progenitor cells and embryonic fibroblasts. We find that lysine 4 and lysine 27 trimethylation effectively discriminates genes that are expressed, poised for expression, or stably repressed, and therefore reflect cell state and lineage potential. Lysine 36 trimethylation marks primary coding and non-coding transcripts, facilitating gene annotation. Trimethylation of lysine 9 and lysine 20 is detected at satellite, telomeric and active long-terminal repeats, and can spread into proximal unique sequences. Lysine 4 and lysine 9 trimethylation marks imprinting control regions. Finally, we show that chromatin state can be read in an allele-specific manner by using single nucleotide polymorphisms. This study provides a framework for the application of comprehensive chromatin profiling towards characterization of diverse mammalian cell populations.
Conflict of interest statement
Author Information
All analyzed data sets can be obtained from www.broad.mit.edu/seq\_platform/chip/. Microarray data have been submitted to the GEO repository under accession number GSE8024. Reprints and permissions information is available from www.nature.com/reprints. The authors declare no competing financial interests.
Figures
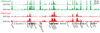
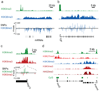
Figure 1. Comparison of ChIP-Seq and ChIP-chip data
Direct comparison of H3K4me3 (green) and H3K27me3 (red) ChIP data across a 300 kb region in mouse ESCs from independent experiments assayed by SMS (absolute fragment counts) or tiling arrays (log p-values for enrichment relative to whole-cell extracts15).
Figure 2. Histone tri-methylation state predicts expression of HCP and LCP promoters
(a) Mammalian promoters can be readily classified into sets with high (HCPs), intermediate (ICPs) or low (LCPs) CpG-content. In ES cells (ESCs), virtually all HCPs are marked by H3K4me3, either alone (green) or in combination with H3K27me3 (yellow). In contrast, most LCPs have neither mark (grey). Few promoters are only enriched for H3K27me3 (red). (b) Tri-methylation states of HCPs and LCPs in NPCs (indicated by colors), conditional on their ESC state (indicated below each bar). HCPs marked by H3K4me3 only in ESCs tend to retain this mark. HCPs marked by H3K4me3 and H3K27me3 tend to lose one or both marks, although some remain bivalent. Small, partially overlapping subsets of LCPs are marked by H3K4me3. (c) Tri-methylation states of HCPs and LCPs in MEFs. (d) Changes in expression levels of HCP genes with H3K4me3 alone (left) or also with H3K27me3 (right) upon differentiation to NPCs. Resolution of bivalent promoters to H3K4me3 is associated with increased expression. Boxplots show median (red bar), 25th and 75th percentile expression levels in ESCs. Whiskers show 2.5th and 97.5th percentiles. Asterisks indicate classes with less than 15 genes. (e) Changes in expression levels of LCP genes with H3K4me3 (left) or no mark (right) upon differentiation to NPCs. Gain of H3K4me3 is associated with increased expression.
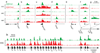
Figure 3. Cell type-specific chromatin marks at promoters
(a) Multiple ‘housekeeping genes’, such as DNA Polymerase mu (Polm), are associated with HCPs marked by H3K4me3 in all cell types. (b) The neural transcription factor gene Olig1 (HCP) is bivalent in ESCs, but resolves to H3K4me3 in NPCs and H3K27me3 in MEFs. (c) The neurogenesis transcription factor gene Neurog1 (HCP) remains bivalent upon differentiation to NPCs, but resolves to H3K27me3 in MEFs. (d) The adipogenesis transcription factor gene _Ppar_-γ (HCP) remains bivalent in MEFs, but loses both marks in NPCs. (e) The neural progenitor marker gene Fabp7 (LCP) is marked by H3K4me3 in NPCs only. (f) The brain and lung expressed transcription factor gene Foxp2 is associated with an HCP that is bivalent in ES cells, but resolves to H3K4me3 in NPCs and remains bivalent in MEFs. (g) Foxp2 also has an LCP marked by H3K4me3 in MEFs only. (h) Multiple, distinct bivalent chromatin marks at the variable region promoters of _Pcdh_-γ. A promoter proximal to the constant region exons (*) is marked by H3K4me3 only.
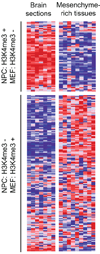
Figure 4. Correlation between chromatin state changes and lineage expression
Relative expression levels across adult mouse brain (frontal and cerebral cortex, substantia nigra, cerebellu, amygdale, hypothalamus, hippocampus) and relatively mesenchyme-rich tissues (bone, white fat, brown fat, trachea, digits, lung, bladder, uterus, umbilical cord) are shown for genes with bivalent chromatin marks in ES cells that retain H3K4me3 in NPCs but lose this mark in MEFs (n=62) or vice versa (n=160). Red, white and blue indicates higher, equal and lower relative expression, respectively.
Figure 5. H3K4me3 and H3K36me3 annotate genes and non-coding RNA transcripts
(a) Foxp1 has two annotated promoters (based on RefSeq and UCSC Known Genes), only one of which shows H3K4me3 in ES cells. The corresponding transcriptional unit is marked by H3K36me3. In MEFs, H3K36me3 extends an additional 500 kb upstream to an H3K4me3 site that appears to reflect an alternate promoter (this site is bivalent in ES cells). (b) H3K36me3 enrichment extends significantly downstream of Sox2. Though highly active in ES cells, Sox2 is flanked by two bivalent CpG islands that may poise it for repression. (c) H3K4me3 and H3K36me3 indicate two highly expressed non-coding RNAs, and (d) the putative primary transcript (dashed line) for a single annotated microRNA.
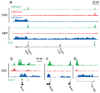
Figure 6. Allele-specific histone methylation and genic H3K9me3/H4K20me3
(a) H3K4me3 and H3K36me3 indicate a primary microRNA transcript in the Dlk1-Dio3 locus. The allele-specificity of this transcript is read out using ChIP-Seq data for hybrid ES cells and a SNP catalogue. The H3K36me3 reads overwhelmingly correspond to maternal 129 alleles, consistent with the known maternal expression of these microRNAs. (b) In contrast, a non-imprinted transcript shows roughly equal proportions of reads assigned to 129 and castaneus alleles. (c) Peg13 is marked by H3K4me3 and H3K9me3 in ES cells; 19 of 21 H3K4me3 reads correspond to the paternal castaneus allele, while 6 of 6 H3K9me3 reads correspond to the maternal 129 allele, consistent with paternal expression of this gene. (d) H3K9 me3 and H4K20me3 enrichment evident at the Polrmt gene may reflect transcriptional interference due to antisense transcription from the 3’ UTR CpG island of Hcn2 (see text).
Comment in
- Genomic biology: the epigenomic era opens.
Baylin SB, Schuebel KE. Baylin SB, et al. Nature. 2007 Aug 2;448(7153):548-9. doi: 10.1038/448548a. Nature. 2007. PMID: 17671496 Free PMC article.
Similar articles
- Chromatin signature of embryonic pluripotency is established during genome activation.
Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Vastenhouw NL, et al. Nature. 2010 Apr 8;464(7290):922-6. doi: 10.1038/nature08866. Epub 2010 Mar 24. Nature. 2010. PMID: 20336069 Free PMC article. - Genomic biology: the epigenomic era opens.
Baylin SB, Schuebel KE. Baylin SB, et al. Nature. 2007 Aug 2;448(7153):548-9. doi: 10.1038/448548a. Nature. 2007. PMID: 17671496 Free PMC article. - Distinct epigenomic landscapes of pluripotent and lineage-committed human cells.
Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Hawkins RD, et al. Cell Stem Cell. 2010 May 7;6(5):479-91. doi: 10.1016/j.stem.2010.03.018. Cell Stem Cell. 2010. PMID: 20452322 Free PMC article. - Chromatin modifiers and remodellers: regulators of cellular differentiation.
Chen T, Dent SY. Chen T, et al. Nat Rev Genet. 2014 Feb;15(2):93-106. doi: 10.1038/nrg3607. Epub 2013 Dec 24. Nat Rev Genet. 2014. PMID: 24366184 Free PMC article. Review. - Epigenetic states in stem cells.
Collas P. Collas P. Biochim Biophys Acta. 2009 Sep;1790(9):900-5. doi: 10.1016/j.bbagen.2008.10.006. Epub 2008 Oct 25. Biochim Biophys Acta. 2009. PMID: 19013220 Review.
Cited by
- Systematic analysis identifies a connection between spatial and genomic variations of chromatin states.
Cao X, Ma T, Fan R, Yuan GC. Cao X, et al. Cell Syst. 2024 Nov 20;15(11):1092-1102.e2. doi: 10.1016/j.cels.2024.10.006. Epub 2024 Nov 13. Cell Syst. 2024. PMID: 39541982 - DNA methylation shapes the Polycomb landscape during the exit from naive pluripotency.
Richard Albert J, Urli T, Monteagudo-Sánchez A, Le Breton A, Sultanova A, David A, Scarpa M, Schulz M, Greenberg MVC. Richard Albert J, et al. Nat Struct Mol Biol. 2024 Oct 24. doi: 10.1038/s41594-024-01405-4. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39448850 - ALYREF enhances breast cancer progression by regulating EZH2.
Jeong SJ, Oh JH, Cho JY. Jeong SJ, et al. Heliyon. 2024 Sep 16;10(19):e37749. doi: 10.1016/j.heliyon.2024.e37749. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39386827 Free PMC article. - scNanoSeq-CUT&Tag: a single-cell long-read CUT&Tag sequencing method for efficient chromatin modification profiling within individual cells.
Li Q, Guo Y, Wu Z, Xu X, Jiang Z, Qi S, Liu Z, Wen L, Tang F. Li Q, et al. Nat Methods. 2024 Nov;21(11):2044-2057. doi: 10.1038/s41592-024-02453-w. Epub 2024 Oct 7. Nat Methods. 2024. PMID: 39375575 - KLF15 suppresses stemness of pancreatic cancer by decreasing USP21-mediated Nanog stability.
Jiang W, Liu L, Wang M, Li X, Zhou T, Hou X, Qiao L, Chen C, Zuo D, Liu J, Ren L. Jiang W, et al. Cell Mol Life Sci. 2024 Oct 5;81(1):417. doi: 10.1007/s00018-024-05442-6. Cell Mol Life Sci. 2024. PMID: 39367978 Free PMC article.
References
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. - PubMed
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. - PubMed
- Mockler TC, et al. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources