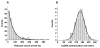Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase - PubMed (original) (raw)
Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase
Daniel S Johnson et al. Cell. 2007.
Abstract
Helicases are molecular motors that separate DNA strands for efficient replication of genomes. We probed the kinetics of individual ring-shaped T7 helicase molecules as they unwound double-stranded DNA (dsDNA) or translocated on single-stranded DNA (ssDNA). A distinctive DNA sequence dependence was observed in the unwinding rate that correlated with the local DNA unzipping energy landscape. The unwinding rate increased approximately 10-fold (approaching the ssDNA translocation rate) when a destabilizing force on the DNA fork junction was increased from 5 to 11 pN. These observations reveal a fundamental difference between the mechanisms of ring-shaped and nonring-shaped helicases. The observed force-velocity and sequence dependence are not consistent with a simple passive unwinding model. However, an active unwinding model fully supports the data even though the helicase on its own does not unwind at its optimal rate. This work offers insights into possible ways helicase activity is enhanced by associated proteins.
Figures
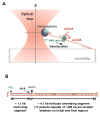
Figure 1. Experimental Configuration
(A) This cartoon illustrates the experimental configuration for the observation of the unwinding of dsDNA by a single T7 helicase (not to scale). One strand of the dsDNA to be unwound was attached to a trapped microsphere via a biotin/streptavidin connection. The other strand was anchored to a microscope coverslip surface via a dig/antidig connection on a dsDNA anchoring segment. The microsphere was held in a feedback-enhanced optical trap so that its position relative to the trap center and the force on it could be measured. Helicase unwinding of dsDNA was monitored as an increase in the ssDNA length. Helicase did not unwind the dsDNA anchoring segment because it prefers two ssDNA regions at a fork junction (Ahnert and Patel, 1997). (B) DNA construct contained both digoxigenin (dig) and biotin labels for binding to the coverglass and microsphere respectively. The dig label was located at one end of the DNA construct and was bound to an anti-digoxigenin coated coverglass. The biotin label was on one strand of the DNA and was bound to a streptavidin-coated microsphere. On the same strand near this biotin label was a nick in the ssDNA. The nick allowed the DNA to be mechanically unwound (unzipped) when the DNA anchor on the coverslip was moved away from the microsphere.
Figure 2. Experimental Method for Measurements of Helicase Translocation along a ssDNA and a Typical Data Set
The experimental procedures were: 1) Mechanically unwind ~ 400 bp of dsDNA at a constant velocity of 1400 bp/s to produce a ssDNA loading region. 2) Maintain the DNA extension until force drops below a threshold indicating helicase unwinding of the DNA fork. 3) Mechanically unwind another ~ 500 bp (Δ_L)_ of dsDNA to generate ssDNA for helicase translocation. 4) Maintain new DNA fork position until force drops again indicating the helicase has caught up with the fork. The blue regions represent the two mechanical unwinding steps and the red regions represent the two helicase unwinding events with intervening time Δ_t_. Helicase translocation velocity on ssDNA was computed as Δ_L_/Δ_t_.
Figure 3. Measurements of Helicase Arrival Time at the Fork Junction and Helicase Translocation Velocity on ssDNA
(A) Measurement of the time it took a helicase to bind to the 400 bp ssDNA loading region and then reach the DNA junction. With the helicase concentration used in our experiments (2 nM monomer), this time followed a single exponential distribution (fit in red) with an average time of ~ 80 s (N = 237). This average helicase arrival time at the junction was much longer than the typical observation time for subsequent ssDNA translocation or DNA unwinding measurements (see Supplementary Discussion). Therefore, it was unlikely that more than one helicase acted on the same DNA template. (B) A histogram of ssDNA helicase translocation rate. Data were pooled from many measurements and the pooled histogram was well fit by a Gaussian distribution with a mean of 322 nt/s and a standard deviation of 62 nt/s.
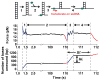
Figure 4. Experimental Method for the Determination of Helicase Unwinding Velocity Under a Constant Force and a Typical Data Set
The experimental procedures were: 1) Mechanically unwind ~ 400 bp of dsDNA to produce a ssDNA loading region. 2) Maintain the DNA extension until force drops indicating helicase is unwinding dsDNA at the junction. 3) Allow force to continue to drop until force reaches a desired value. 4) Maintain constant force while helicase is unwinding dsDNA. The blue (red) region of the data represents the mechanical (helicase) unwinding.
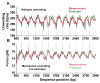
Figure 5. Sequence Dependence of Helicase Unwinding Velocity
(A) Instantaneous velocity of helicase along dsDNA template positions under 11 pN force. The measured curve was obtained by averaging 32 single traces. The measured sequence-dependent velocity agreed well with that predicted from an active model. (B) Measured and predicted dsDNA mechanical unwinding force on the same DNA sequence. Unzipping force anti-correlates with the helicase unwinding rate in Figure 5A.
Figure 6. Force-velocity Measurements During Helicase Unwinding
(A) Example traces of helicase unwinding of dsDNA at various forces. Unwinding velocity varied dramatically with the applied force on ssDNA. (B) Measured force-velocity relation and its comparison with predictions based on various models. Measured force-velocity relation deviated significantly from a simple passive model at step sizes (δ) 1, 2, or 3 bp; larger δ only made the deviation larger. In contrast, an active model with δ = 2 bp and destabilization energy Δ_G_d = 1.2 k_B_T per bp over an 6 bp range agrees well with the measurements. Also marked on the plot are the ssDNA translocation rate _k_0 and the minimum (critical) DNA mechanical unwinding force _F_c for the given sequence. Error bars indicate standard errors of the means.
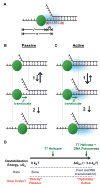
Figure 7. Cartoon of Passive and Active Unwinding Mechanisms
(A) A sketch illustrating the notation used for modeling the helicase movement towards a fork junction. l is the number of nucleotides between the 5′ end of the ssDNA and the helicase. m is the number nucleotides of the ssDNA that are open between the helicase and the junction. Therefore, n = l + m is the total number of nucleotides from the end of the ssDNA to the junction. M defines the range of interactions between the helicase and the junction in nt. (B) Illustration of the passive unwinding mechanism. In this model the DNA fork thermally fluctuates between dsDNA and ssDNA states (step 1). When the amount of ssDNA between the helicase and the junction is greater than or equal to the helicase step size (δ) the helicase may forward translocate (step 2). (C) Illustration of the active unwinding mechanism. In this model the helicase destabilizes a region (the light blue cloud) of dsDNA near the junction (step 1). This makes the junction more likely to be open so that the helicase is able to step forward (step 2) more frequently. (D) A cartoon illustrating the degree of activeness in DNA unwinding by helicase. When Δ_G_d = 0, the helicase may be considered “strictly” passive, and when Δ_G_d ~ Δ_G_GC (~ 3.4 k_B_T, GC base pairing energy), “optimally active”. For T7 helicase Δ_G_d ~ 1–2 k_B_T.
Comment in
- Need for speed: mechanical regulation of a replicative helicase.
Ha T. Ha T. Cell. 2007 Jun 29;129(7):1249-50. doi: 10.1016/j.cell.2007.06.007. Cell. 2007. PMID: 17604712 Review.
Similar articles
- Need for speed: mechanical regulation of a replicative helicase.
Ha T. Ha T. Cell. 2007 Jun 29;129(7):1249-50. doi: 10.1016/j.cell.2007.06.007. Cell. 2007. PMID: 17604712 Review. - Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase.
Patel SS, Pandey M, Nandakumar D. Patel SS, et al. Curr Opin Chem Biol. 2011 Oct;15(5):595-605. doi: 10.1016/j.cbpa.2011.08.003. Epub 2011 Aug 22. Curr Opin Chem Biol. 2011. PMID: 21865075 Free PMC article. Review. - The DNA-unwinding mechanism of the ring helicase of bacteriophage T7.
Jeong YJ, Levin MK, Patel SS. Jeong YJ, et al. Proc Natl Acad Sci U S A. 2004 May 11;101(19):7264-9. doi: 10.1073/pnas.0400372101. Epub 2004 May 3. Proc Natl Acad Sci U S A. 2004. PMID: 15123793 Free PMC article. - DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase.
Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. Stano NM, et al. Nature. 2005 May 19;435(7040):370-3. doi: 10.1038/nature03615. Nature. 2005. PMID: 15902262 Free PMC article. - On translocation mechanism of ring-shaped helicase along single-stranded DNA.
Xie P. Xie P. Biochim Biophys Acta. 2007 Jun;1774(6):737-48. doi: 10.1016/j.bbapap.2007.04.002. Epub 2007 Apr 13. Biochim Biophys Acta. 2007. PMID: 17499029
Cited by
- Pif1 is a force-regulated helicase.
Li JH, Lin WX, Zhang B, Nong DG, Ju HP, Ma JB, Xu CH, Ye FF, Xi XG, Li M, Lu Y, Dou SX. Li JH, et al. Nucleic Acids Res. 2016 May 19;44(9):4330-9. doi: 10.1093/nar/gkw295. Epub 2016 Apr 20. Nucleic Acids Res. 2016. PMID: 27098034 Free PMC article. - Active DNA unwinding dynamics during processive DNA replication.
Morin JA, Cao FJ, Lázaro JM, Arias-Gonzalez JR, Valpuesta JM, Carrascosa JL, Salas M, Ibarra B. Morin JA, et al. Proc Natl Acad Sci U S A. 2012 May 22;109(21):8115-20. doi: 10.1073/pnas.1204759109. Epub 2012 May 9. Proc Natl Acad Sci U S A. 2012. PMID: 22573817 Free PMC article. - Different mechanisms for translocation by monomeric and hexameric helicases.
Gao Y, Yang W. Gao Y, et al. Curr Opin Struct Biol. 2020 Apr;61:25-32. doi: 10.1016/j.sbi.2019.10.003. Epub 2019 Nov 26. Curr Opin Struct Biol. 2020. PMID: 31783299 Free PMC article. Review. - Synergy between RecBCD subunits is essential for efficient DNA unwinding.
Zananiri R, Malik O, Rudnizky S, Gaydar V, Kreiserman R, Henn A, Kaplan A. Zananiri R, et al. Elife. 2019 Jan 2;8:e40836. doi: 10.7554/eLife.40836. Elife. 2019. PMID: 30601118 Free PMC article. - Ring-shaped replicative helicase encircles double-stranded DNA during unwinding.
Joo S, Chung BH, Lee M, Ha TH. Joo S, et al. Nucleic Acids Res. 2019 Dec 2;47(21):11344-11354. doi: 10.1093/nar/gkz893. Nucleic Acids Res. 2019. PMID: 31665506 Free PMC article.
References
- Ahnert P, Patel SS. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of forked DNA substrate. J Biol Chem. 1997;272:32267–32273. - PubMed
- Amaratunga M, Lohman TM. Escherichia coli rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;27:6815–6820. - PubMed
- Betterton MD, Jülicher F. A motor that makes its own track: helicase unwinding of DNA. Phys Rev Lett. 2003;91:258103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM059849-05/GM/NIGMS NIH HHS/United States
- R01 GM059849/GM/NIGMS NIH HHS/United States
- R01 GM059849-04/GM/NIGMS NIH HHS/United States
- R01 GM055310/GM/NIGMS NIH HHS/United States
- R01 GM059849-07/GM/NIGMS NIH HHS/United States
- R01 GM059849-09/GM/NIGMS NIH HHS/United States
- GM55310/GM/NIGMS NIH HHS/United States
- R01 GM059849-06A1/GM/NIGMS NIH HHS/United States
- R01 GM059849-08/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources