Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer - PubMed (original) (raw)
. 2007 Aug;6(8):2993-3002.
doi: 10.1021/pr060629m. Epub 2007 Jul 4.
Affiliations
- PMID: 17608509
- PMCID: PMC2584611
- DOI: 10.1021/pr060629m
Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer
Flaubert Mbeunkui et al. J Proteome Res. 2007 Aug.
Abstract
Proteins secreted (the secretome) from cancer cells are potentially useful as biomarkers of the disease. Using LC-MS/MS, the secreted proteomes from a series of isogenic breast cancer cell lines varying in aggressiveness were analyzed by mass spectrometry: nontumorigenic MCF10A, premalignant/tumorigenic MCF10AT, tumorigenic/locally invasive MCF10 DCIS.com, and tumorigenic/metastatic MCF 10CA cl. D. Proteomes were obtained from conditioned serum-free media, partially fractionated using a small reverse phase C2 column, and digested with trypsin for analysis by LC-MS/MS, using a method previously shown to give highly enriched secreted proteomes (Mbeunkui et al. J. Proteome Res. 2006, 5, 899-906). The search files produced from five analyses (three separate preparations) were combined for database searching (Mascot) which produced a list of over 250 proteins from each cell line. The aim was to discover highly secreted proteins which changed significantly in abundance corresponding with aggressiveness. The most apparent changes were observed for alpha-1-antichymotrypsin and galectin-3-binding protein which were highly secreted proteins from MCF10 DCIS.com and MCF10CA cl. D, yet undetected in the MCF10A and MCF10AT cell lines. Other proteins showing increasing abundance in the more aggressive cell lines included alpha-1-antitrypsin, cathepsin D, and lysyl oxidase. The S100 proteins, often associated with metastasis, showed variable changes in abundance. While the cytosolic proteins were low (e.g., actin and tubulin), there was significant secretion of proteins often associated with the cytoplasm. These proteins were all predicted as products of nonclassical secretion (SecretomeP, Center for Biological Sequence Analysis). The LC-MS/MS results were verified for five selected proteins by western blot analysis, and the relevance of other significant proteins is discussed. Comparisons with two other aggressive breast cancer cell lines are included. The protein with consistent association with aggressiveness in all lines, and in unrelated cancer cells, was the galectin-3-binding protein which has been associated with breast, prostate, and colon cancer earlier, supporting the approach and findings. This analysis of an isogenic series of cell lines suggests the potential usefulness of the secretome for identifying prospective markers for the early detection and aggressiveness/progression of cancer.
Figures
Figure 1
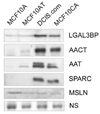
The profile of the secreted proteome changes as the cells become progressively more aggressive. Conditioned serum free media from the cell lines was concentrated and the total protein estimated. Eight micrograms of total protein was resolved by SDS-PAGE and immunoblotted for LGAL3BP (galectin-3-binding protein), AACT (alpha-1-chymoantitrypsin), AAT (alpha-1-antitrypsin), SPARC (osteonectin) and MSLN (mesothelin). NS represents a non-specific band used to show equal loading.
Figure 2
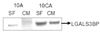
The profile of the secreted proteome remains unchanged even in the absence of any serum in the growth medium for 16–18 hours. Conditioned media (containing 5% FBS (CM) as well as serum free (SF) media) from the cell lines was concentrated eight-fold. Albumin was depleted from the serum-containing samples. Eight micrograms of total protein was resolved by SDS-PAGE and immunoblotted for galectin-3-binding protein (LGALS3BP). 10A corresponds to MCF10A cells and 10CA to MCF10CA cl. D cells.
Scheme 1
View of the overall strategy for secreted proteome comparisons by LC-MS/MS with Mascot searching and verification by western blotting.
Similar articles
- Mass spectrometry (LC-MS/MS) identified proteomic biosignatures of breast cancer in proximal fluid.
Whelan SA, He J, Lu M, Souda P, Saxton RE, Faull KF, Whitelegge JP, Chang HR. Whelan SA, et al. J Proteome Res. 2012 Oct 5;11(10):5034-45. doi: 10.1021/pr300606e. Epub 2012 Sep 20. J Proteome Res. 2012. PMID: 22934887 Free PMC article. - Proteomic Analysis of Urine to Identify Breast Cancer Biomarker Candidates Using a Label-Free LC-MS/MS Approach.
Beretov J, Wasinger VC, Millar EK, Schwartz P, Graham PH, Li Y. Beretov J, et al. PLoS One. 2015 Nov 6;10(11):e0141876. doi: 10.1371/journal.pone.0141876. eCollection 2015. PLoS One. 2015. PMID: 26544852 Free PMC article. - Comparative membrane proteomics analyses of breast cancer cell lines to understand the molecular mechanism of breast cancer brain metastasis.
Peng W, Zhang Y, Zhu R, Mechref Y. Peng W, et al. Electrophoresis. 2017 Sep;38(17):2124-2134. doi: 10.1002/elps.201700027. Epub 2017 Jul 5. Electrophoresis. 2017. PMID: 28523741 - The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context.
Schaaij-Visser TB, de Wit M, Lam SW, Jiménez CR. Schaaij-Visser TB, et al. Biochim Biophys Acta. 2013 Nov;1834(11):2242-58. doi: 10.1016/j.bbapap.2013.01.029. Epub 2013 Jan 31. Biochim Biophys Acta. 2013. PMID: 23376433 Review. - Secretome Proteomic Approaches for Biomarker Discovery: An Update on Colorectal Cancer.
Cevenini A, Orrù S, Imperlini E. Cevenini A, et al. Medicina (Kaunas). 2020 Aug 31;56(9):443. doi: 10.3390/medicina56090443. Medicina (Kaunas). 2020. PMID: 32878319 Free PMC article. Review.
Cited by
- Global secretome analysis identifies novel mediators of bone metastasis.
Blanco MA, LeRoy G, Khan Z, Alečković M, Zee BM, Garcia BA, Kang Y. Blanco MA, et al. Cell Res. 2012 Sep;22(9):1339-55. doi: 10.1038/cr.2012.89. Epub 2012 Jun 12. Cell Res. 2012. PMID: 22688892 Free PMC article. - Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers.
Whelan SA, Lu M, He J, Yan W, Saxton RE, Faull KF, Whitelegge JP, Chang HR. Whelan SA, et al. J Proteome Res. 2009 Aug;8(8):4151-60. doi: 10.1021/pr900322g. J Proteome Res. 2009. PMID: 19522481 Free PMC article. - Proteomic identification of multitasking proteins in unexpected locations complicates drug targeting.
Butler GS, Overall CM. Butler GS, et al. Nat Rev Drug Discov. 2009 Dec;8(12):935-48. doi: 10.1038/nrd2945. Nat Rev Drug Discov. 2009. PMID: 19949400 Review. - Differential Expression of Key Signaling Proteins in MCF10 Cell Lines, a Human Breast Cancer Progression Model.
So JY, Lee HJ, Kramata P, Minden A, Suh N. So JY, et al. Mol Cell Pharmacol. 2012 Jan 1;4(1):31-40. Mol Cell Pharmacol. 2012. PMID: 24558516 Free PMC article. - Phenotypic and Molecular Characterization of MCF10DCIS and SUM Breast Cancer Cell Lines.
Barnabas N, Cohen D. Barnabas N, et al. Int J Breast Cancer. 2013;2013:872743. doi: 10.1155/2013/872743. Epub 2013 Jan 16. Int J Breast Cancer. 2013. PMID: 23401782 Free PMC article.
References
- Flagg EW, et al. Validation of the American Cancer Society Cancer Prevention Study II Nutrition Survey Cohort Food Frequency Questionnaire. Epidemiology. 2000;11(4):462–468. - PubMed
- Ruoslahti E. Fibronectin and its integrin receptors in cancer. Adv Cancer Res. 1999;76:1–20. - PubMed
- Mbeunkui F, Fodstad O, Pannell LK. Secretory protein enrichment and analysis: an optimized approach applied on cancer cell lines using 2D LC-MS/MS. J Proteome Res. 2006;5(4):899–906. - PubMed
- Hunter T. Oncoprotein networks. Cell. 1997;88(3):333–346. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous