A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast - PubMed (original) (raw)
A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast
Vibha Taneja et al. Mol Cell. 2007.
Abstract
Prions are self-propagating, infectious aggregates of misfolded proteins. The mammalian prion, PrP(Sc), causes fatal neurodegenerative disorders. Fungi also have prions. While yeast prions depend upon glutamine/asparagine (Q/N)-rich regions, the Podospora anserina HET-s and PrP prion proteins lack such sequences. Nonetheless, we show that the HET-s prion domain fused to GFP propagates as a prion in yeast. Analogously to native yeast prions, transient overexpression of the HET-s fusion induces ring-like aggregates that propagate in daughter cells as cytoplasmically inherited, detergent-resistant dot aggregates. Efficient dot propagation, but not ring formation, is dependent upon the Hsp104 chaperone. The yeast prion [PIN(+)] enhances HET-s ring formation, suggesting that prions with and without Q/N-rich regions interact. Finally, HET-s aggregates propagated in yeast are infectious when introduced into Podospora. Taken together, these results demonstrate prion propagation in a truly foreign host. Since yeast can host non-Q/N-rich prions, such native yeast prions may exist.
Figures
Figure 1. Aggregates of HET-s (PrD)-GFP in yeast
The HET-s(PrD)-GFP fusion construct was expressed in a [_pin_−][_psi_−] strain. (A) The expression levels of Het-s(PrD)-GFP fusion on 0.05% gal and 2% gal media in log (OD600nm ~0.3) and stationary phase (OD600nm ~1.2). Arrows indicate the Het-s(PrD)-GFP fusion. (B-J) Cells were examined under a fluorescent microscope after 48 hours of growth at 30°C. Diffuse fluorescence (B) was observed when cells were grown on 0.05% gal where the fusion is expressed at a ‘low level’. Aggregates of different shapes were formed when cells were grown on 2% gal, which induced ‘high expression’ of the fusion. Rings were (C) smooth, (D) branched, (E-F) twisted, (G) theta-like, or (H) internal. In addition, there were dots (I), and the T266P mutant of PrD-GFP showed no aggregates (J).
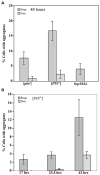
Figure 2. Quantitative measure of HET-s(PrD)-GFP aggregation in yeast
Yeast [_psi_−] strains containing the HET-s(PrD)-GFP plasmid were induced to form aggregates on 2% gal. The percentage of cells with aggregates in three (A), and four (B), independent transformants was determined by examining 300–600 cells for each. Error bars indicate standard deviations. (A) The induction of Het-s(PrD)-GFP rings and dots after 48 hours of growth in 2% gal in strains with the indicated genotypes. (B) The appearance of rings and dots when a [PIN+] strain is grown in 2% gal for the indicated periods of time.
Figure 3. HET-s(PrD)-GFP ring-like aggregates give rise to dots
A. [PIN+][_psi_−] yeast with HET-s(PrD)-GFP was induced to form rings on 2% gal (left panel) for 48 hours. The cells were then passed again to 2% gal (bottom panel) or 3-times on 0.05% gal (right panel). B. As a control, the cells were grown only on 0.05% gal. C. A liquid culture in 2% gal was briefly sonicated and plated as isolated cells on solid 2% gal. Microcolonies of 2 to 8-cells were observed after 12 hours. Cells with dots mainly correspond to daughters or granddaughters of cells with rings. Since rings can sometimes only be visualized in their entirety by varying the focal plane, cells with rings are marked with an arrowhead for clarity.
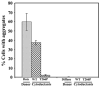
Figure 4. HET-s(PrD)-GFP aggregates formed in yeast are cytoducible
[Het-s]y or [het-s]y [PIN+] HSP104 donor strains grown on 0.05% gal after induction on 2% gal and with, respectively Het-s aggregates (dots) or diffuse fluorescence, were cytoduced into a [_rho_−] [_pin_−] [het-s]y HSP104 cyh2R recipient carrying either pHET-s(PrD)-GFP (WT) or mutant pT266P-GFP plasmids. Recipients and cytoductants were grown only on 0.05% gal. Data is the average of five and three independent cytoductions (300–500 cells for each) respectively for [Het-s]y and [het-s]y donor strains.
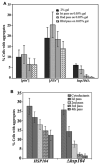
Figure 5. Hsp104 is required for the maintenance of HET-s(PrD)-GFP aggregates
A. The maintenance of HET-s aggregates in the indicated strains was monitored on three serial passes of the cells on 0.05% gal after induction of HET-s(PrD) on 2% gal. The percentage of cells with aggregates was determined after 36–40 hours of growth on inducing media and subsequent passes on 0.05% gal in four transformants (300–500 cells each) for each strain. B. HSP104 donor strain with Het-s dots was mated with both HSP104 and _hsp104_Δ strains. The cytoductants obtained on SG-Trp+0.05% gal+Cyh were passed serially four times on SR-Trp+0.05% gal. The percentage of Het-s dots in eight independent cytoductants and subsequent passes were determined after 36 hours of growth on the respective media.
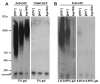
Figure 6. HET-s(PrD)-GFP aggregates formed in yeast are sarkosyl resistant
Precleared yeast lysates normalized for total protein and treated with 2% sarkosyl at room temperature for 10 min were resolved on 1.5% agarose gels and probed with anti-GFP antibody. Stained chicken pectoralis muscle extract (Kim and Keller, 2002) provided molecular weight markers. The position of Het-s(PrD)-GFP monomer is shown, although the monomer does not reliably transfer on agarose gels. Indeed, we established that neither [PIN+], _hsp104_Δ or T266P altered the total level of Het-s(PrD)-GFP in cells by probing blots of acrylamide gels with anti-GFP antibody (Figure S1A) A. Left panel: The HET-s aggregates formed in the indicated strains following growth in 2% gal were broken into sarkosyl resistant subparticles. Right panel represents strains expressing the T266P-GFP fusion in 2% gal. B. Left panel: HSP104 cells with HET-s(PrD)-GFP aggregates obtained in 2% gal even after two serial passes in 0.05% gal gave rise to sarkosyl resistant subparticles. Subparticles were lost in _hsp104_Δ. Right panel represents strains after two serial passes in 0.05% gal without prior growth in 2% gal.
Similar articles
- Analyzing the birth and propagation of two distinct prions, [PSI+] and Het-s, in yeast.
Mathur V, Taneja V, Sun Y, Liebman SW. Mathur V, et al. Mol Biol Cell. 2010 May 1;21(9):1449-61. doi: 10.1091/mbc.e09-11-0927. Epub 2010 Mar 10. Mol Biol Cell. 2010. PMID: 20219972 Free PMC article. - Role of Hsp104 in the propagation and inheritance of the [Het-s] prion.
Malato L, Dos Reis S, Benkemoun L, Sabaté R, Saupe SJ. Malato L, et al. Mol Biol Cell. 2007 Dec;18(12):4803-12. doi: 10.1091/mbc.e07-07-0657. Epub 2007 Sep 19. Mol Biol Cell. 2007. PMID: 17881723 Free PMC article. - The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion.
Patel BK, Gavin-Smyth J, Liebman SW. Patel BK, et al. Nat Cell Biol. 2009 Mar;11(3):344-9. doi: 10.1038/ncb1843. Epub 2009 Feb 15. Nat Cell Biol. 2009. PMID: 19219034 Free PMC article. - Infectious fold and amyloid propagation in Podospora anserina.
Maddelein ML. Maddelein ML. Prion. 2007 Jan-Mar;1(1):44-7. doi: 10.4161/pri.1.1.4083. Epub 2007 Jan 28. Prion. 2007. PMID: 19164904 Free PMC article. Review. - Yeast prions: evolution of the prion concept.
Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T, Engel A, McCann L, Kryndushkin D. Wickner RB, et al. Prion. 2007 Apr-Jun;1(2):94-100. doi: 10.4161/pri.1.2.4664. Epub 2007 Apr 28. Prion. 2007. PMID: 19164928 Free PMC article. Review.
Cited by
- How Do Yeast Cells Contend with Prions?
Wickner RB, Edskes HK, Son M, Wu S, Niznikiewicz M. Wickner RB, et al. Int J Mol Sci. 2020 Jul 3;21(13):4742. doi: 10.3390/ijms21134742. Int J Mol Sci. 2020. PMID: 32635197 Free PMC article. Review. - Curcumin prevents formation of polyglutamine aggregates by inhibiting Vps36, a component of the ESCRT-II complex.
Verma M, Sharma A, Naidu S, Bhadra AK, Kukreti R, Taneja V. Verma M, et al. PLoS One. 2012;7(8):e42923. doi: 10.1371/journal.pone.0042923. Epub 2012 Aug 7. PLoS One. 2012. PMID: 22880132 Free PMC article. - Interdependence of amyloid formation in yeast: implications for polyglutamine disorders and biological functions.
Urakov VN, Vishnevskaya AB, Alexandrov IM, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Urakov VN, et al. Prion. 2010 Jan-Mar;4(1):45-52. doi: 10.4161/pri.4.1.11074. Epub 2010 Jan 18. Prion. 2010. PMID: 20118659 Free PMC article. - Reciprocal efficiency of RNQ1 and polyglutamine detoxification in the cytosol and nucleus.
Douglas PM, Summers DW, Ren HY, Cyr DM. Douglas PM, et al. Mol Biol Cell. 2009 Oct;20(19):4162-73. doi: 10.1091/mbc.e09-02-0170. Epub 2009 Aug 5. Mol Biol Cell. 2009. PMID: 19656852 Free PMC article. - Prion propagation can occur in a prokaryote and requires the ClpB chaperone.
Yuan AH, Garrity SJ, Nako E, Hochschild A. Yuan AH, et al. Elife. 2014 Aug 13;3:e02949. doi: 10.7554/eLife.02949. Elife. 2014. PMID: 25122461 Free PMC article.
References
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. - PubMed
- Baron T. Mouse models of prion disease transmission. Trends Mol Med. 2002;8:495–500. - PubMed
- Baxa U, Cassese T, Kajava AV, Steven AC. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv Protein Chem. 2006;73:125–180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials