Transcriptional control of brown fat determination by PRDM16 - PubMed (original) (raw)
Transcriptional control of brown fat determination by PRDM16
Patrick Seale et al. Cell Metab. 2007 Jul.
Abstract
Brown fat cells are specialized to dissipate energy and can counteract obesity; however, the transcriptional basis of their determination is largely unknown. We show here that the zinc-finger protein PRDM16 is highly enriched in brown fat cells compared to white fat cells. When expressed in white fat cell progenitors, PRDM16 activates a robust brown fat phenotype including induction of PGC-1alpha, UCP1, and type 2 deiodinase (Dio2) expression and a remarkable increase in uncoupled respiration. Transgenic expression of PRDM16 at physiological levels in white fat depots stimulates the formation of brown fat cells. Depletion of PRDM16 through shRNA expression in brown fat cells causes a near total loss of the brown characteristics. PRDM16 activates brown fat cell identity at least in part by simultaneously activating PGC-1alpha and PGC-1beta through direct protein binding. These data indicate that PRDM16 can control the determination of brown fat fate.
Figures
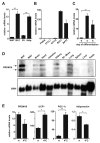
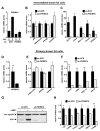
Figure 1. PRDM16 is expressed selectively in brown fat cells
(A) Real-time PCR analysis of PRDM16 mRNA expression in BAT relative to WAT; and its expression in the adipocyte fraction (Adip.) compared to the stromal vascular fraction (SV) of BAT (n= 6, mean ± SD). (B) PRDM16 mRNA expression in adipocytes from immortalized BAT cell lines (BAT1 and BAT2) and three WAT cell lines (3T3-F442A, 3T3-L1, C3H-10T1/2) (n=3–5 samples per cell line, mean ± SD). (C) PRDM16 mRNA levels during the differentiation of immortalized BAT cell lines (n=3 for each of 2 separate cell lines, mean ± SD). (D) Northern blot analysis of PRDM16 mRNA and control, 36B4 mRNA, in adult mouse tissues. (E) Expression of PRDM16, UCP1, PGC-1α and adiponectin in BAT after a 4°C cold exposure of mice for 4 hours (n=5 mice per group, mean ± SD) (rt: room temperature). * p < 0.05; ** p < 0.01.
Figure 2. PRDM16 expression induces the gene program of brown fat cells
(A) Oil-red-O staining of mature adipocytes (day 6) from PPARγ-deficient cells expressing retroviral PPARγ2 and either retroviral- PRDM16 or vector control (ctl). These adipocyte cultures were analyzed by real-time PCR for their expression of: differentiation markers common to WAT and BAT (A); brown fat cell-selective genes (as indicated) (B); brown fat- thermogenic genes (UCP1, deiodinase-d2 (dio-2), PGC-1α) with and without cAMP treatment (C); mitochondrial components (as indicated) (D); and white fat cell-selective markers (psat1, serpin3ak, resistin) (E). (n=3, mean ± SD). * p < 0.05; ** p < 0.01
Figure 3. PRDM16 stimulates mitochondrial biogenesis and uncoupled respiration
(A) Representative transmission electron micrographs of fibroblasts and mature adipocytes (day 6) from PPARγ-deficient cells expressing retroviral PPARγ2 and either retroviral- PRDM16 or vector control (ctl). (B) Comparison of mitochondrial volume densities from cells depicted in (A) (n= >20 micrographs per group, mean ± SD). (C, D) Total mitochondrial oxygen consumption and uncoupled respiration in mature adipocytes expressing either PRDM16 or control vector under basal conditions (n=4, mean ± SD) (C), or after stimulation with a cAMP analog for 12 hours (n=4, mean ± SD) (D). L: lipid droplet. * p < 0.05; ** p < 0.01
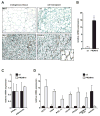
Figure 4. Differentiation of PRDM16-expressing cells into brown fat in vivo
(A) Immunohistochemistry for cidea protein expression in endogenous WAT and BAT, and in ectopic subcutaneous fat pads formed from fibroblasts expressing PPARγ2 and either vector control (ctl) or PRDM16. (B–D) 8-weeks after transplantation, ectopic fat pads were analyzed by real-time PCR for expression of: PRDM16 (B); differentiation markers common to white and brown fat (PPARγ and adiponectin) (C); brown fat-selective genes (UCP1, cidea, PGC-1α, elovl3, PPAR-α, endogenous PRDM16 (-3′UTR) and the white fat selective marker, resistin (D). (n=10 mice per cell line, mean ± SE). * p < 0.05; ** p < 0.01
Figure 5. PRDM16 activates PGC-1α and PGC-1β via direct binding
(A) The transcriptional activity of the -2 kb region of PGC-1α in response to PRDM16 or vector control expression in brown fat preadipocytes (n=3, mean ± SD). (B) PGC-1α promoter activity in response to PRDM16 or vector expression in PGC-1α-deficient cells (n=3, mean ± SD). (C) The transcriptional activity of Gal4-DNA binding domain (DBD) fusion proteins containing PGC-1α, PGC-1β, or PRC in response to PRDM16 or vector expression (n=3, mean ± SD). (D) Flag-PRDM16 and its associated proteins were immunoprecipitated from brown fat preadipocytes and analyzed by Western blot to detect PGC-1α and PGC-1β (E) GST fusion proteins containing different regions of PGC-1α were incubated with 35S- labeled PRDM16 protein. ERR-α was used to demonstrate binding to the 1–190 and 200–350 regions of PGC-1α. (F) PGC-1α and PRDM16 were co-precipitated from cos7 cells that had been transfected with HA-PGC-1α and either wildtype (WT) or R998Q mutant PRDM16. The input was 2% of the cell lysate used for immunoprecipitation. (G) WT or R998Q mutant PRDM16 were expressed with PPARγ2 in PPARγ−/− fibroblasts. After differentiation into adipocytes (day 6), real-time PCR was used to measure the mRNA expression of: brown fat-selective genes (as indicated); and resistin, a white fat selective gene (n=3, mean ± SD). * p < 0.05; ** p < 0.01
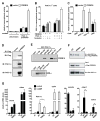
Figure 6. Knockdown of PRDM16 in brown fat cells ablates their brown fat characteristics
(A) PRDM16 mRNA levels in immortalized brown fat cells expressing shRNA targeted to PRDM16 or a scrambled (SCR) control- shRNA before (day 0) and after their differentiation (day 5) into adipocytes. (B) Gene expression in brown fat cells (day 5) expressing sh-PRDM16 or sh-SCR including: markers common to white and brown fat cells (aP2, PPARγ, adiponectin) and resistin, a white fat cell selective gene. (C) The differentiation-linked mRNA induction (day 0 to day 5) of brown fat- selective genes (as indicated) in sh-PRDM16 and sh-SCR expressing cells. (D–F) Gene expression in adipocytes (day 6) from sh-PRDM16 and sh-SCR expressing primary brown preadipocytes including mRNA levels of: PRDM16 (D); adiponectin, PPARγ and resistin (E); brown fat- selective genes (as indicated) (F). (G) Western blot analysis of UCP1 protein levels in primary brown fat cells expressing sh-PRDM16 or sh-SCR control with and without cAMP treatment. (H) mRNA levels of various mitochondrial components in adipocytes from sh-PRDM16 and sh-SCR expressing primary brown preadipocytes. (n= 3–5, mean ± SD). * p < 0.05; ** p < 0.01
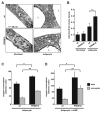
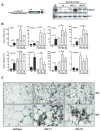
Figure 7. Transgenic expression of PRDM16 in WAT depots induces the formation of BAT cells
(A) The fat-specific aP2 promoter/enhancer was used to express PRDM16 in WAT depots. Western blot analysis for PRDM16 protein expression in: non-transgenic, wildtype (wt) BAT; wt WAT; and WAT from two strains of aP2-PRDM16 transgenic mice (aP2-T1 and aP2-T2). POL-II protein expression was used to control for loading. (B) Expression of BAT-selective genes (as indicated) and resistin in WAT from wildtype (wt) and aP2-T1 transgenic mice. This gene set was also measured in WAT from wt, aP2-T1 and aP2-T2 mice that had been treated with CL 316, 243 (n= 7–10 mice per group, mean ± SE). (C) Immunohistochemistry for UCP1 protein (brown stain) in sections of WAT from wt and transgenic mice (T1 and T2) after treatment with CL 316, 243. * p < 0.05; ** p < 0.01
Similar articles
- ERRγ enhances UCP1 expression and fatty acid oxidation in brown adipocytes.
Dixen K, Basse AL, Murholm M, Isidor MS, Hansen LH, Petersen MC, Madsen L, Petrovic N, Nedergaard J, Quistorff B, Hansen JB. Dixen K, et al. Obesity (Silver Spring). 2013 Mar;21(3):516-24. doi: 10.1002/oby.20067. Obesity (Silver Spring). 2013. PMID: 23404793 - Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation.
Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Uldry M, et al. Cell Metab. 2006 May;3(5):333-41. doi: 10.1016/j.cmet.2006.04.002. Cell Metab. 2006. PMID: 16679291 - Two key temporally distinguishable molecular and cellular components of white adipose tissue browning during cold acclimation.
Jankovic A, Golic I, Markelic M, Stancic A, Otasevic V, Buzadzic B, Korac A, Korac B. Jankovic A, et al. J Physiol. 2015 Aug 1;593(15):3267-80. doi: 10.1113/JP270805. Epub 2015 Jul 14. J Physiol. 2015. PMID: 26096127 Free PMC article. - Role of PRDM16 in the activation of brown fat programming. Relevance to the development of obesity.
Becerril S, Gómez-Ambrosi J, Martín M, Moncada R, Sesma P, Burrell MA, Frühbeck G. Becerril S, et al. Histol Histopathol. 2013 Nov;28(11):1411-25. doi: 10.14670/HH-28.1411. Epub 2013 Jun 17. Histol Histopathol. 2013. PMID: 23771475 Review. - Positive and negative control of Ucp1 gene transcription and the role of β-adrenergic signaling networks.
Collins S, Yehuda-Shnaidman E, Wang H. Collins S, et al. Int J Obes (Lond). 2010 Oct;34 Suppl 1:S28-33. doi: 10.1038/ijo.2010.180. Int J Obes (Lond). 2010. PMID: 20935662 Review.
Cited by
- Natural Bioactive Compounds as Potential Browning Agents in White Adipose Tissue.
Choi Y, Yu L. Choi Y, et al. Pharm Res. 2021 Apr;38(4):549-567. doi: 10.1007/s11095-021-03027-7. Epub 2021 Mar 30. Pharm Res. 2021. PMID: 33783666 Free PMC article. Review. - PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1.
Iida S, Chen W, Nakadai T, Ohkuma Y, Roeder RG. Iida S, et al. Genes Dev. 2015 Feb 1;29(3):308-21. doi: 10.1101/gad.252809.114. Genes Dev. 2015. PMID: 25644605 Free PMC article. - Human adipose dynamics and metabolic health.
Feng B, Zhang T, Xu H. Feng B, et al. Ann N Y Acad Sci. 2013 Apr;1281(1):160-77. doi: 10.1111/nyas.12009. Epub 2013 Jan 14. Ann N Y Acad Sci. 2013. PMID: 23317303 Free PMC article. Review. - CPEB2-activated Prdm16 translation promotes brown adipocyte function and prevents obesity.
Lu WH, Chen HF, King PC, Peng C, Huang YS. Lu WH, et al. Mol Metab. 2024 Nov;89:102034. doi: 10.1016/j.molmet.2024.102034. Epub 2024 Sep 19. Mol Metab. 2024. PMID: 39305947 Free PMC article. - The Engrailed-1 Gene Stimulates Brown Adipogenesis.
Zhang C, Weng Y, Shi F, Jin W. Zhang C, et al. Stem Cells Int. 2016;2016:7369491. doi: 10.1155/2016/7369491. Epub 2016 Apr 11. Stem Cells Int. 2016. PMID: 27148369 Free PMC article.
References
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. - PubMed
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. - PubMed
- Barbera MJ, Schluter A, Pedraza N, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J Biol Chem. 2001;276:1486–1493. - PubMed
- Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK031405/DK/NIDDK NIH HHS/United States
- R37 DK031405/DK/NIDDK NIH HHS/United States
- R37 DK031405-25/DK/NIDDK NIH HHS/United States
- DK31405-24/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials