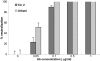Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies - PubMed (original) (raw)
. 2007 Jul 17;104(29):12123-8.
doi: 10.1073/pnas.0701000104. Epub 2007 Jul 9.
Samitabh Chakraborti, Yuxian He, Anjeanette Roberts, Tim Sheahan, Xiaodong Xiao, Lisa E Hensley, Ponraj Prabakaran, Barry Rockx, Igor A Sidorov, Davide Corti, Leatrice Vogel, Yang Feng, Jae-Ouk Kim, Lin-Fa Wang, Ralph Baric, Antonio Lanzavecchia, Kristopher M Curtis, Gary J Nabel, Kanta Subbarao, Shibo Jiang, Dimiter S Dimitrov
Affiliations
- PMID: 17620608
- PMCID: PMC1924550
- DOI: 10.1073/pnas.0701000104
Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies
Zhongyu Zhu et al. Proc Natl Acad Sci U S A. 2007.
Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) caused a worldwide epidemic in late 2002/early 2003 and a second outbreak in the winter of 2003/2004 by an independent animal-to-human transmission. The GD03 strain, which was isolated from an index patient of the second outbreak, was reported to resist neutralization by the human monoclonal antibodies (hmAbs) 80R and S3.1, which can potently neutralize isolates from the first outbreak. Here we report that two hmAbs, m396 and S230.15, potently neutralized GD03 and representative isolates from the first SARS outbreak (Urbani, Tor2) and from palm civets (SZ3, SZ16). These antibodies also protected mice challenged with the Urbani or recombinant viruses bearing the GD03 and SZ16 spike (S) glycoproteins. Both antibodies competed with the SARS-CoV receptor, ACE2, for binding to the receptor-binding domain (RBD), suggesting a mechanism of neutralization that involves interference with the SARS-CoV-ACE2 interaction. Two putative hot-spot residues in the RBD (Ile-489 and Tyr-491) were identified within the SARS-CoV spike that likely contribute to most of the m396-binding energy. Residues Ile-489 and Tyr-491 are highly conserved within the SARS-CoV spike, indicating a possible mechanism of the m396 cross-reactivity. Sequence analysis and mutagenesis data show that m396 might neutralize all zoonotic and epidemic SARS-CoV isolates with known sequences, except strains derived from bats. These antibodies exhibit cross-reactivity against isolates from the two SARS outbreaks and palm civets and could have potential applications for diagnosis, prophylaxis, and treatment of SARS-CoV infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
M396 potently neutralizes viruses pseudotyped with S glycoproteins from the Tor2 and GD03 isolates. HIVs pseudotyped with the S glycoprotein from Tor2 and GD03 isolates were incubated with IgG1 m396 for 1 h before infection. Luciferase activities in target cells were measured, and the percent neutralization was calculated. All experiments were performed in duplicate or triplicate, and two experiments in different days were performed with essentially identical results. Bars indicate SE.
Fig. 2.
Potent neutralization of replication-competent virus by m396. Tor2 and Urbani isolates were incubated with IgG1 m396 for 1 h at 37°C before infection. After incubation, the percent neutralization was determined by plaque reduction assay in Vero E6 cells (in duplicate) compared with untreated controls. Bars indicate SE.
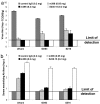
Fig. 3.
Potent neutralization of replication-competent recombinant SARS-CoV in mice after antibody administration. (a) BALB/c mice, 8 weeks old, were injected i.p with a control monoclonal antibody at 200 μg per mouse; m396 at 50 or 200 μg per mouse; or S230.15 at 200 μg per mouse. Twenty-four hours after antibody administration, mice were bled to evaluate antibody levels in serum and then challenged intranasally with 105 TCID50 of the respective recombinant SARS-CoV (icUrbani, icGD03, or icSZ16-K479N (SZ16). Virus titers in the lung, determined 2 days after challenge, are expressed as log10 TCID50 per g of lung tissue (limit of detection ≤101.5 TCID50 per g of lung tissue). (b) Serum-neutralizing antibodies were measured against specific challenge viruses by microneutralization assays. The log10-transformed reciprocal dilution at which 50% neutralization occurred is indicated (limit of detection <8 or 100.9). Bars indicate SE.
Fig. 4.
Schematic representation of the SARS-CoV neutralization mechanism. Competition of the antibody (Ab, Fab m396) with the receptor (ACE2) for binding to the receptor-binding site (RBS) of the RBD of the SARS-CoV S glycoprotein is shown. The protruding portion of the antibody epitope (in violet) is also a major portion of the ACE2 receptor-binding site.
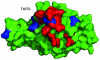
Fig. 5.
Amino acid residues that are different in GD03 compared with Urbani (in blue) are located outside the m396 epitope (in red). The antibody contact residues are shown in red on the surface of the RBD crystal structure determined in our previous study (36).
Fig. 6.
Analysis of available SARS-CoV sequences and mutagenesis data. M396 is likely to neutralize all isolates with known sequences. Percentage variability is calculated as the ratio of the number of isolates with a specific mutation to the total number of sequences (72) multiplied by 100. Mutations in SARS-CoV RBD sequences are shown in blue. Residues critical for binding to m396 are shown in red. The RBD residues that are in contact with both m396 and ACE2 are underlined. Mutations of noncontact residues that lead to significant decrease of the m396 binding are denoted by an asterisk.
Similar articles
- Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein.
He Y, Li J, Li W, Lustigman S, Farzan M, Jiang S. He Y, et al. J Immunol. 2006 May 15;176(10):6085-92. doi: 10.4049/jimmunol.176.10.6085. J Immunol. 2006. PMID: 16670317 - Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants.
ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E, Preiser W, Doerr HW, Chow VT, de Kruif J, Peiris JS, Goudsmit J. ter Meulen J, et al. PLoS Med. 2006 Jul;3(7):e237. doi: 10.1371/journal.pmed.0030237. PLoS Med. 2006. PMID: 16796401 Free PMC article. - Pathways of cross-species transmission of synthetically reconstructed zoonotic severe acute respiratory syndrome coronavirus.
Sheahan T, Rockx B, Donaldson E, Corti D, Baric R. Sheahan T, et al. J Virol. 2008 Sep;82(17):8721-32. doi: 10.1128/JVI.00818-08. Epub 2008 Jun 25. J Virol. 2008. PMID: 18579604 Free PMC article. - A review of studies on animal reservoirs of the SARS coronavirus.
Shi Z, Hu Z. Shi Z, et al. Virus Res. 2008 Apr;133(1):74-87. doi: 10.1016/j.virusres.2007.03.012. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451830 Free PMC article. Review. - Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential.
Coughlin MM, Prabhakar BS. Coughlin MM, et al. Rev Med Virol. 2012 Jan;22(1):2-17. doi: 10.1002/rmv.706. Epub 2011 Sep 8. Rev Med Virol. 2012. PMID: 21905149 Free PMC article. Review.
Cited by
- Convalescent plasma anti-SARS-CoV-2 spike protein ectodomain and receptor-binding domain IgG correlate with virus neutralization.
Salazar E, Kuchipudi SV, Christensen PA, Eagar T, Yi X, Zhao P, Jin Z, Long SW, Olsen RJ, Chen J, Castillo B, Leveque C, Towers D, Lavinder J, Gollihar J, Cardona J, Ippolito G, Nissly R, Bird I, Greenawalt D, Rossi RM, Gontu A, Srinivasan S, Poojary I, Cattadori IM, Hudson PJ, Josleyn NM, Prugar L, Huie K, Herbert A, Bernard DW, Dye JM, Kapur V, Musser JM. Salazar E, et al. J Clin Invest. 2020 Dec 1;130(12):6728-6738. doi: 10.1172/JCI141206. J Clin Invest. 2020. PMID: 32910806 Free PMC article. Clinical Trial. - Nanoluciferase-based cell fusion assay for rapid and high-throughput assessment of SARS-CoV-2-neutralizing antibodies in patient samples.
Meyrath M, Szpakowska M, Plesseria JM, Domingues O, Langlet J, Weber B, Krüger R, Ollert M, Chevigné A; CON-VINCE Consortium. Meyrath M, et al. Methods Enzymol. 2022;675:351-381. doi: 10.1016/bs.mie.2022.07.015. Epub 2022 Sep 9. Methods Enzymol. 2022. PMID: 36220277 Free PMC article. - Antibodies at work in the time of severe acute respiratory syndrome coronavirus 2.
Sajna KV, Kamat S. Sajna KV, et al. Cytotherapy. 2021 Feb;23(2):101-110. doi: 10.1016/j.jcyt.2020.08.009. Epub 2020 Aug 31. Cytotherapy. 2021. PMID: 32988772 Free PMC article. Review. - De novo design and Rosetta-based assessment of high-affinity antibody variable regions (Fv) against the SARS-CoV-2 spike receptor binding domain (RBD).
Boorla VS, Chowdhury R, Ramasubramanian R, Ameglio B, Frick R, Gray JJ, Maranas CD. Boorla VS, et al. Proteins. 2023 Feb;91(2):196-208. doi: 10.1002/prot.26422. Epub 2022 Oct 8. Proteins. 2023. PMID: 36111441 Free PMC article. - An integrated drug repurposing strategy for the rapid identification of potential SARS-CoV-2 viral inhibitors.
Trezza A, Iovinelli D, Santucci A, Prischi F, Spiga O. Trezza A, et al. Sci Rep. 2020 Aug 17;10(1):13866. doi: 10.1038/s41598-020-70863-9. Sci Rep. 2020. PMID: 32807895 Free PMC article.
References
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, et al. N Engl J Med. 2003;348:1953–1966. - PubMed
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, et al. N Engl J Med. 2003;348:1967–1976. - PubMed
- Holmes KV. N Engl J Med. 2003;348:1948–1951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI059443/AI/NIAID NIH HHS/United States
- AI059136/AI/NIAID NIH HHS/United States
- N01 CO012400/CA/NCI NIH HHS/United States
- R01 AI059136/AI/NIAID NIH HHS/United States
- N01-CO-12400/CO/NCI NIH HHS/United States
- AI059434/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous