Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington's disease - PubMed (original) (raw)
Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington's disease
Xiaoyun Liu et al. J Proteome Res. 2007 Aug.
Abstract
Liquid chromatography (LC) coupled with mass spectrometry (MS) and database assignment methods have been used to conduct a large-scale proteome survey of the R6/2 mouse model of Huntington's disease (HD). Although the neuropathological mechanisms of HD are not known, the mutant huntingtin gene that causes the disease is thought to alter gene transcription, leading to a cascade of neurotoxic events. In this report, we have focused on characterizing changes in the brain proteome associated with HD pathophysiology. Differences in the relative abundances of proteins (R6/2 compared to wild type) in brain tissue from the striatum and cortex, two primary loci of dysfunction in HD, were assessed by using a label-free approach based on calibrations to internal standards. In total, assignments were made for approximately 400 proteins. A set of criteria was used to establish 160 high confidence assignments, approximately 30% of which appear to show differences in expression relative to wild type (WT) animals. Many of the proteins that were differentially expressed are known to be associated with neurotransmission and likely play key roles in HD etiology. This study is the first to report that the majority of differentially expressed proteins in the striatum are up-regulated, while the majority of the expressed proteins in the cortex are down-regulated. The differentially expressed proteins identified in this proteomic screen may be potential biomarkers and drug targets for HD and may further our understanding of the disease pathology.
Figures
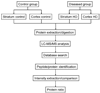
Figure 1
Schematic flow diagram of the experimental approach used for the comparative proteomics analysis of mouse brain samples.
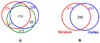
Figure 2
Venn diagrams showing the protein overlap in different experiments. Part A shows a Venn diagram representing the number of proteins observed for each of the triplicate runs of the diseased striatum sample, as well as the protein overlap. Part B shows a Venn diagram summary of the proteins identified from striatum and cortex by LC-MS/MS analysis. In total, 957 and 967 peptides have been observed in striatum and cortex respectively, corresponding to 417 and 403 protein identifications, respectively.
Figure 3
Part A shows plots of extracted ion chromatograms for the peptide ion [GITWGEETLMEYLENPKK+2H]2+ obtained from triplicate runs with the left and center plots acquired in LC-MS/MS mode and the right plot acquired in LC-MS mode. Part B shows extracted ion chromatograms for [TSVNVVGDSFGAGIVYHLSK+2H]2+ obtained from triplicate runs with the left and center plots acquired in LC-MS/MS mode and the right plot acquired in LC-MS mode. We note that for both plots only a partial view of the ion chromatogram (with the peak of interest) is shown such that three peaks of the same ion can be displayed side by side within a single plot for better visual comparison.
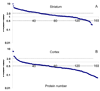
Figure 4
Observed protein abundance ratios in striatum (A) and cortex (B). Positive ratios have been calculated by dividing intensity of R6/2 samples by that of WT samples and plotted on a logarithmic scale. Proteins (represented by a series of numbers) are sorted by descending order of abundance ratios. See text for discussion.
Figure 5
Collision-induced dissociation spectrum leading to the assignment of the [TSVNVVGDSFGAGIVYHLSK+2H]2+ peptide ion from GLT1. A MASCOT score of 135 is obtained from these data. See text for details.
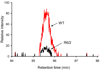
Figure 6
Extracted ion chromatograms of the GLT1 peptide ion [TSVNVVGDSFGAGIVYHLSK+2H]2+ from the WT (denoted by red line) and the R6/2 (denoted by black line) samples. See text for details.
Similar articles
- Reduced Expression of Foxp1 as a Contributing Factor in Huntington's Disease.
Louis Sam Titus ASC, Yusuff T, Cassar M, Thomas E, Kretzschmar D, D'Mello SR. Louis Sam Titus ASC, et al. J Neurosci. 2017 Jul 5;37(27):6575-6587. doi: 10.1523/JNEUROSCI.3612-16.2017. Epub 2017 May 26. J Neurosci. 2017. PMID: 28550168 Free PMC article. - Differential proteomic and genomic profiling of mouse striatal cell model of Huntington's disease and control; probable implications to the disease biology.
Choudhury KR, Das S, Bhattacharyya NP. Choudhury KR, et al. J Proteomics. 2016 Jan 30;132:155-66. doi: 10.1016/j.jprot.2015.11.007. Epub 2015 Nov 12. J Proteomics. 2016. PMID: 26581643 - Mass Spectrometry Analysis of Wild-Type and Knock-in Q140/Q140 Huntington's Disease Mouse Brains Reveals Changes in Glycerophospholipids Including Alterations in Phosphatidic Acid and Lyso-Phosphatidic Acid.
Vodicka P, Mo S, Tousley A, Green KM, Sapp E, Iuliano M, Sadri-Vakili G, Shaffer SA, Aronin N, DiFiglia M, Kegel-Gleason KB. Vodicka P, et al. J Huntingtons Dis. 2015;4(2):187-201. doi: 10.3233/JHD-150149. J Huntingtons Dis. 2015. PMID: 26397899 - Brain Cholesterol Synthesis and Metabolism is Progressively Disturbed in the R6/1 Mouse Model of Huntington's Disease: A Targeted GC-MS/MS Sterol Analysis.
Kreilaus F, Spiro AS, Hannan AJ, Garner B, Jenner AM. Kreilaus F, et al. J Huntingtons Dis. 2015;4(4):305-18. doi: 10.3233/JHD-150170. J Huntingtons Dis. 2015. PMID: 26639223 - Genetic manipulations of mutant huntingtin in mice: new insights into Huntington's disease pathogenesis.
Lee CY, Cantle JP, Yang XW. Lee CY, et al. FEBS J. 2013 Sep;280(18):4382-94. doi: 10.1111/febs.12418. Epub 2013 Jul 31. FEBS J. 2013. PMID: 23829302 Free PMC article. Review.
Cited by
- The heat shock response in neurons and astroglia and its role in neurodegenerative diseases.
San Gil R, Ooi L, Yerbury JJ, Ecroyd H. San Gil R, et al. Mol Neurodegener. 2017 Sep 18;12(1):65. doi: 10.1186/s13024-017-0208-6. Mol Neurodegener. 2017. PMID: 28923065 Free PMC article. Review. - Proteome response to the panneural expression of human wild-type alpha-synuclein: a Drosophila model of Parkinson's disease.
Xun Z, Kaufman TC, Clemmer DE. Xun Z, et al. J Proteome Res. 2008 Sep;7(9):3911-21. doi: 10.1021/pr800207h. Epub 2008 Aug 7. J Proteome Res. 2008. PMID: 18683964 Free PMC article. - Structural separations by ion mobility-MS for glycomics and glycoproteomics.
Fenn LS, McLean JA. Fenn LS, et al. Methods Mol Biol. 2013;951:171-94. doi: 10.1007/978-1-62703-146-2_12. Methods Mol Biol. 2013. PMID: 23296531 Free PMC article. - Mutant huntingtin disrupts mitochondrial proteostasis by interacting with TIM23.
Yablonska S, Ganesan V, Ferrando LM, Kim J, Pyzel A, Baranova OV, Khattar NK, Larkin TM, Baranov SV, Chen N, Strohlein CE, Stevens DA, Wang X, Chang YF, Schurdak ME, Carlisle DL, Minden JS, Friedlander RM. Yablonska S, et al. Proc Natl Acad Sci U S A. 2019 Aug 13;116(33):16593-16602. doi: 10.1073/pnas.1904101116. Epub 2019 Jul 25. Proc Natl Acad Sci U S A. 2019. PMID: 31346086 Free PMC article. - Taming the Huntington's Disease Proteome: What Have We Learned?
Seeley C, Kegel-Gleason KB. Seeley C, et al. J Huntingtons Dis. 2021;10(2):239-257. doi: 10.3233/JHD-200465. J Huntingtons Dis. 2021. PMID: 33998547 Free PMC article. Review.
References
- Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded an unstable on Huntington's disease chromosomes. Cell. 1993;72:971–993. - PubMed
- Bates GP. Lancet. 2003;361:1642–1644. - PubMed
- Browne SE, Beal MF. Neurochem. Res. 2004;29:531–546. - PubMed
- Bence NF, Sampat RM, Kopito RR. Science. 2001;292:1552–1555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG024547/AG/NIA NIH HHS/United States
- R01 NS035663/NS/NINDS NIH HHS/United States
- R01 AG 024547/AG/NIA NIH HHS/United States
- R01 NS 35663/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical