A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing - PubMed (original) (raw)
A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing
Debopriya Das et al. Nucleic Acids Res. 2007.
Abstract
Correlation of motif occurrences with gene expression intensity is an effective strategy for elucidating transcriptional cis-regulatory logic. Here we demonstrate that this approach can also identify cis-regulatory elements for alternative pre-mRNA splicing. Using data from a human exon microarray, we identified 56 cassette exons that exhibited higher transcript-normalized expression in muscle than in other normal adult tissues. Intron sequences flanking these exons were then analyzed to identify candidate regulatory motifs for muscle-specific alternative splicing. Correlation of motif parameters with gene-normalized exon expression levels was examined using linear regression and linear splines on RNA words and degenerate weight matrices, respectively. Our unbiased analysis uncovered multiple candidate regulatory motifs for muscle-specific splicing, many of which are phylogenetically conserved among vertebrate genomes. The most prominent downstream motifs were binding sites for Fox1- and CELF-related splicing factors, and a branchpoint-like element acuaac; pyrimidine-rich elements resembling PTB-binding sites were most significant in upstream introns. Intriguingly, our systematic study indicates a paucity of novel muscle-specific elements that are dominant in short proximal intronic regions. We propose that Fox and CELF proteins play major roles in enforcing the muscle-specific alternative splicing program, facilitating expression of unique isoforms of cytoskeletal proteins critical to muscle cell function.
Figures
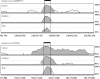
Figure 1.
Validation of microarray predictions of muscle-enriched alternative exons. RT-PCR confirmation of muscle-enriched alternative exon expression. Amplifications were performed using primers in the flanking constitutive exons. RNA sources used for amplification by lane number: 1, brain; 2, kidney; 3, liver; 4, stomach; 5, bone marrow; 6, testis; 7, heart; 8, skeletal muscle. Arrowheads indicate positions of the alternative exon inclusion products that are most enriched in heart and skeletal muscle.
Figure 2.
Conservation of intron sequences flanking muscle-enriched exons. Representative VISTA genome alignments of exon and flanking intron sequences from the mouse, chicken and frog genomes with the prototypical muscle-enriched exon from human. Exon boundaries are indicated by vertical lines. Shaded regions indicate sequences that exceed 75% identity, while curves above baseline indicate regions with >50% identity to the human sequence.
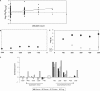
Figure 3.
Correlation with exon expression for the
ugcaug
regulatory element. (A) Linear fit between ratios of gene-normalized exon expression levels and counts of
ugcaug
in 200 nt of downstream intronic sequence across 356 exons (56 muscle-specific exons and 300 randomly selected exons) (P = 4.6_E_–06). (B) Dependence of correlational _P_-values of
ugcaug
count with distance. Filled circles indicate correlation with total motif count, while unfilled boxes indicate correlation with bin-wise count (bin size = 100 nt). E.g. for 300 nt, the filled circle reflects the strength of correlation with the count in 1–300 nt of intron, while the unfilled box reflects the correlation with the count in 201–300 nt of intron. Minus sign indicates upstream intron. (C) Contrast scores of
ugcaug
in upstream and downstream introns of human, mouse, chicken and frog. (‘E’ in Figure 3B and 3C indicates position of the exon).
Figure 4.
Phylogenetic conservation of regulatory motifs in the proximal intron sequences. Bar graphs show the over-representation of the indicated _cis_-regulatory motifs in the proximal intron sequences for muscle-enriched exons in four vertebrate species. (A) Enrichment of selected regulatory motifs in 1-kb flanking intron regions. The highest abundance of
ugcaug, ugugug
and
acuaac
elements is consistently within the proximal downstream region of ∼200 nt, while
cucucu
and
ucuu
elements were enriched in the proximal upstream intron. The representative hnRNP A1-binding site
uaggg
was not over-represented near muscle-enriched exons. (B) Analysis of the putative CELF-binding site
ugc
in 0.5-kb flanking introns.
ugc
is enriched in the D50 region of all four species. Vertical axis, contrast score, i.e. difference in motif frequency between muscle datasets and control datasets of constitutive exons, occurrences/nt × 103; horizontal axis, nt range relative to the alternative exon; E indicates position of the muscle-enriched exon.
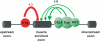
Figure 5.
A candidate model showing splicing factors implicated in regulation of conserved muscle-enriched alternative exons. Based on the conserved distribution of splicing factor binding sites across multiple vertebrate orders, the positive correlation with muscle-specific splicing and the high absolute abundance of
ugcaug
motifs, Fox proteins are proposed to play a major role in promoting inclusion of muscle-enriched exons. The distribution of binding motifs among individual introns suggests that CELF proteins and by KH-type splicing factor(s) function independently in some cases, and together with Fox proteins in others, to specify muscle-enriched splicing. In contrast, the enrichment of candidate PTB-binding sites in the proximal upstream intron suggests a role in preventing inappropriate inclusion of muscle-specific exons in other cell types.
Similar articles
- Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities.
Nakahata S, Kawamoto S. Nakahata S, et al. Nucleic Acids Res. 2005 Apr 11;33(7):2078-89. doi: 10.1093/nar/gki338. Print 2005. Nucleic Acids Res. 2005. PMID: 15824060 Free PMC article. - Intronic alternative splicing regulators identified by comparative genomics in nematodes.
Kabat JL, Barberan-Soler S, McKenna P, Clawson H, Farrer T, Zahler AM. Kabat JL, et al. PLoS Comput Biol. 2006 Jul 14;2(7):e86. doi: 10.1371/journal.pcbi.0020086. Epub 2006 Jun 5. PLoS Comput Biol. 2006. PMID: 16839192 Free PMC article. - Systematic characterization of short intronic splicing-regulatory elements in SMN2 pre-mRNA.
Gao Y, Lin KT, Jiang T, Yang Y, Rahman MA, Gong S, Bai J, Wang L, Sun J, Sheng L, Krainer AR, Hua Y. Gao Y, et al. Nucleic Acids Res. 2022 Jan 25;50(2):731-749. doi: 10.1093/nar/gkab1280. Nucleic Acids Res. 2022. PMID: 35018432 Free PMC article. - Fox-1 family of RNA-binding proteins.
Kuroyanagi H. Kuroyanagi H. Cell Mol Life Sci. 2009 Dec;66(24):3895-907. doi: 10.1007/s00018-009-0120-5. Cell Mol Life Sci. 2009. PMID: 19688295 Free PMC article. Review. - Splicing in action: assessing disease causing sequence changes.
Baralle D, Baralle M. Baralle D, et al. J Med Genet. 2005 Oct;42(10):737-48. doi: 10.1136/jmg.2004.029538. J Med Genet. 2005. PMID: 16199547 Free PMC article. Review.
Cited by
- SpliceTransformer predicts tissue-specific splicing linked to human diseases.
You N, Liu C, Gu Y, Wang R, Jia H, Zhang T, Jiang S, Shi J, Chen M, Guan MX, Sun S, Pei S, Liu Z, Shen N. You N, et al. Nat Commun. 2024 Oct 23;15(1):9129. doi: 10.1038/s41467-024-53088-6. Nat Commun. 2024. PMID: 39443442 Free PMC article. - The Role of Alternative Splicing Factors hnRNP G and Fox-2 in the Progression and Prognosis of Esophageal Cancer.
Zheng Y, Niu X, Xue W, Li L, Geng Q, Fan Z, Zhao J. Zheng Y, et al. Dis Markers. 2022 Nov 23;2022:3043737. doi: 10.1155/2022/3043737. eCollection 2022. Dis Markers. 2022. PMID: 36466711 Free PMC article. - Sensitive period-regulating genetic pathways and exposure to adversity shape risk for depression.
Zhu Y, Wang MJ, Crawford KM, Ramírez-Tapia JC, Lussier AA, Davis KA, de Leeuw C, Takesian AE; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; Hensch TK, Smoller JW, Dunn EC. Zhu Y, et al. Neuropsychopharmacology. 2022 Jan;47(2):497-506. doi: 10.1038/s41386-021-01172-6. Epub 2021 Oct 23. Neuropsychopharmacology. 2022. PMID: 34689167 Free PMC article. - The regulatory genome of the malaria vector Anopheles gambiae: integrating chromatin accessibility and gene expression.
Ruiz JL, Ranford-Cartwright LC, Gómez-Díaz E. Ruiz JL, et al. NAR Genom Bioinform. 2021 Jan 20;3(1):lqaa113. doi: 10.1093/nargab/lqaa113. eCollection 2021 Mar. NAR Genom Bioinform. 2021. PMID: 33987532 Free PMC article. - The evolution of hemocyanin genes in Tectipleura: a multitude of conserved introns in highly diverse gastropods.
Schäfer GG, Pedrini-Martha V, Jackson DJ, Dallinger R, Lieb B. Schäfer GG, et al. BMC Ecol Evol. 2021 Mar 4;21(1):36. doi: 10.1186/s12862-021-01763-3. BMC Ecol Evol. 2021. PMID: 33663373 Free PMC article.
References
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. - PubMed
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. - PubMed
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. - PubMed