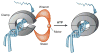Cell cycle regulation of DNA replication - PubMed (original) (raw)
Review
Cell cycle regulation of DNA replication
R A Sclafani et al. Annu Rev Genet. 2007.
Abstract
Eukaryotic DNA replication is regulated to ensure all chromosomes replicate once and only once per cell cycle. Replication begins at many origins scattered along each chromosome. Except for budding yeast, origins are not defined DNA sequences and probably are inherited by epigenetic mechanisms. Initiation at origins occurs throughout the S phase according to a temporal program that is important in regulating gene expression during development. Most replication proteins are conserved in evolution in eukaryotes and archaea, but not in bacteria. However, the mechanism of initiation is conserved and consists of origin recognition, assembly of prereplication (pre-RC) initiative complexes, helicase activation, and replisome loading. Cell cycle regulation by protein phosphorylation ensures that pre-RC assembly can only occur in G1 phase, whereas helicase activation and loading can only occur in S phase. Checkpoint regulation maintains high fidelity by stabilizing replication forks and preventing cell cycle progression during replication stress or damage.
Figures
Figure 1
Models of the regulation of DNA replication. (a) In coliphage λ replication, origin is recognized by λO protein, then λP protein loads hexameric DnaB helicase. λP protein is removed by DnaJ-K protein, which activates the helicase and allows replisome to replicate the DNA. (b) In eukaryotic DNA replication, origin is recognized by ORC, then Cdc6 and Cdt1 protein load the hexameric MCM helicase to form the “licensed” (L) pre-RC in G1 phase (L = 1, A = 0). Geminin inhibits Cdt1 and pre-RC formation. CDK and DDK become active in late G1, activate (A) the MCM helicase and load on the replisome that contains the DNA polymerases. In addition, CDK inhibits any further licensing (L = 0, A = 1). Toward this end, CDK phosphorylates Sld2 and Sld3 proteins and DDK phosphorylates MCM proteins, which “pushes out” the “A” domain of Mcm5.
Figure 2
Structures of ORC/Cdc6 and DNA helicases. (a) Ribbon diagram of the atomic structure of the N-terminal fragment of a single archaeal Mth-MCM subunit (b) rotated 90°. A, B, and C domains are indicated. Arrow indicates the position of the P62 residue (76). (c) EM reconstruction of yeast Orc/Cdc6 complex with ORC in blue (258) (d) EM reconstruction of the full-length archaeal double hexameric Mth-MCM complex (96). (e) Ribbon diagram of the atomic structure of a single hexamer of SV40 T antigen (161). (f) Space-filling diagram of the atomic structure of the N-terminal fragment of a single hexamer of the archaeal Mth-MCM complex (left) and a cut side-view (right) with two subunits removed for clarity; blue indicates positively charged amino acids, red indicates negatively charged amino acids (76).
Figure 3
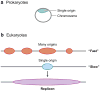
Regulation of DNA replication by origin usage. (a) Prokaryotes have a single origin on a circular chromosome (above). (b) In eukaryotes, multiple origins are found on a single chromosome. When replication is “fast,” many origins are used, whereas only one origin is used in this region when replication is “slow”. Replication proceeds bidirectionally from an origin to form a replicon (below).
Figure 4
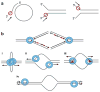
The clamp loading mechanism. Clamp loader (orange) opens clamp using the energy from ATP hydrolysis. Clamp loader is composed of “stator,” “wrench,” and “motor” functions. Clamp loader fixes clamp onto the “stator” while opening the clamp with the “wrench” and “motor.” Open clamp is bound to DNA and then closed. Adapted from (52).
Figure 5
Helicase substrates and models. (a) In vitro helicase substrates that are used frequently have small ssDNA (50 bp) annealed to ssDNA circle (5 kb) with nonhomologous 3′ tail. Helicases (red circle) such as SV40 T antigen or Mcm4/6/7 complex translocate 3′ to 5′ on the tail to unwind DNA and release oligonucleotide from the larger circle. Other substrates used resemble replication forks that are produced by annealing small ssDNA oligonucleotides with nonhomologous ends. Helicases can translocate 3′ to 5′ as above or 5′ to 3′ (DnaB). (b) A single hexameric helicase is depicted as a ring (blue) at the ends of a conventionally drawn replication fork. Lagging strand Okazaki fragments are shown with RNA (red) primers at their 5′ ends. (i) The SV40 T antigen model (161) is made by putting the two rings together forming a loop. In this model, the DNA is pumped into the channel of the double hexamer and then extruded out the holes in the outside C-terminal domains (Figure 2_d_). (ii) In the “pump-in-ring” model, each single hexamer translocates on a different strand of DNA (127). (iii) In the “ploughshare” model, the ploughshare (red) acts as a wedge and keeps the ssDNA unwound as it emerges from behind each single hexamer (271). (iv) In the “rotary pump” model, different single hexamers twist the DNA at a distance resulting in topological strain and unwinding in the center (151).
Figure 6
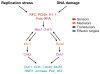
DNA replication and damage checkpoint regulation. Replication stress or blockage or DNA damage induces activation of a signal transduction pathway of many different proteins. The proteins are in different classes indicated as sensors, mediators, transducers, and effector targets. For example, if DNA replication is blocked, ssDNA coated by RPA sends a signal to activate Mec1 protein kinase. Mec1 binds to Mrc1, which amplifies the signal by binding to and activating Chk2 (Rad53) protein kinase. Chk2, in turn, inhibits Cdc45 helicase activation and loading of the replisome.
Similar articles
- Mechanism of chromosomal DNA replication initiation and replication fork stabilization in eukaryotes.
Wu L, Liu Y, Kong D. Wu L, et al. Sci China Life Sci. 2014 May;57(5):482-7. doi: 10.1007/s11427-014-4631-4. Epub 2014 Apr 4. Sci China Life Sci. 2014. PMID: 24699916 Review. - Factors affecting the diversity of DNA replication licensing control in eukaryotes.
Drury LS, Diffley JF. Drury LS, et al. Curr Biol. 2009 Mar 24;19(6):530-5. doi: 10.1016/j.cub.2009.02.034. Epub 2009 Mar 12. Curr Biol. 2009. PMID: 19285403 - Kinetic modelling of DNA replication initiation in budding yeast.
Barberis M, Spiesser TW, Klipp E. Barberis M, et al. Genome Inform. 2010;24:1-20. Genome Inform. 2010. PMID: 22081585 - DNA replication origins, ORC/DNA interaction, and assembly of pre-replication complex in eukaryotes.
Sun J, Kong D. Sun J, et al. Acta Biochim Biophys Sin (Shanghai). 2010 Jul;42(7):433-9. doi: 10.1093/abbs/gmq048. Epub 2010 Jun 9. Acta Biochim Biophys Sin (Shanghai). 2010. PMID: 20705581 Review. - Mechanisms involved in regulating DNA replication origins during the cell cycle and in response to DNA damage.
Early A, Drury LS, Diffley JF. Early A, et al. Philos Trans R Soc Lond B Biol Sci. 2004 Jan 29;359(1441):31-8. doi: 10.1098/rstb.2003.1362. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15065654 Free PMC article. Review.
Cited by
- Hypoxic reactivation of Kaposi's sarcoma associated herpesvirus.
Singh RK, Torne AS, Robertson ES. Singh RK, et al. Cell Insight. 2024 Sep 7;3(6):100200. doi: 10.1016/j.cellin.2024.100200. eCollection 2024 Dec. Cell Insight. 2024. PMID: 39391006 Free PMC article. Review. - ALTMAN: A Novel Method for Cell Cycle Analysis.
Wang Z, Wang T, Chen X, Lv L, Luo Y, Gu W. Wang Z, et al. ACS Omega. 2024 Jul 22;9(36):37780-37788. doi: 10.1021/acsomega.4c03653. eCollection 2024 Sep 10. ACS Omega. 2024. PMID: 39281911 Free PMC article. - Multifaceted role of the DNA replication protein MCM10 in maintaining genome stability and its implication in human diseases.
Ahmed SMQ, Sasikumar J, Laha S, Das SP. Ahmed SMQ, et al. Cancer Metastasis Rev. 2024 Dec;43(4):1353-1371. doi: 10.1007/s10555-024-10209-3. Epub 2024 Sep 6. Cancer Metastasis Rev. 2024. PMID: 39240414 Review. - Selectively advantageous instability in biotic and pre-biotic systems and implications for evolution and aging.
Tower J. Tower J. Front Aging. 2024 May 16;5:1376060. doi: 10.3389/fragi.2024.1376060. eCollection 2024. Front Aging. 2024. PMID: 38818026 Free PMC article. Review. - The Crk4-Cyc4 complex regulates G2/M transition in Toxoplasma gondii.
Hawkins LM, Wang C, Chaput D, Batra M, Marsilia C, Awshah D, Suvorova ES. Hawkins LM, et al. EMBO J. 2024 Jun;43(11):2094-2126. doi: 10.1038/s44318-024-00095-4. Epub 2024 Apr 10. EMBO J. 2024. PMID: 38600241 Free PMC article.
References
- Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–79. - PubMed
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–76. - PubMed
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, et al. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–3. - PubMed
- Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005;7:831–7. - PubMed
- Alexandrov AI, Botchan MR, Cozzarelli NR. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. The active form of the helicase. J Biol Chem. 2002;277:44886–277. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM035078-15A2/GM/NIGMS NIH HHS/United States
- R01 GM035078/GM/NIGMS NIH HHS/United States
- GM 35078/GM/NIGMS NIH HHS/United States
- R01 GM035078-16/GM/NIGMS NIH HHS/United States
- R01 GM035078-17/GM/NIGMS NIH HHS/United States
- R01 GM035078-18/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases