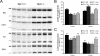Cholesterol accumulation is associated with lysosomal dysfunction and autophagic stress in Npc1 -/- mouse brain - PubMed (original) (raw)
Cholesterol accumulation is associated with lysosomal dysfunction and autophagic stress in Npc1 -/- mouse brain
Guanghong Liao et al. Am J Pathol. 2007 Sep.
Abstract
Niemann-Pick type C (NPC) disease is an autosomal recessive disorder caused by mutations of NPC1 and NPC2 genes. Progressive neurodegeneration that accompanies NPC is fatal, but the underlying mechanisms are still poorly understood. In the present study, we characterized the association of autophagic-lysosomal dysfunction with cholesterol accumulation in Npc1(-/-) mice during postnatal development. Brain levels of lysosomal cathepsin D were significantly higher in mutant than in wild-type mice. Increases in cathepsin D occurred first in neurons and later in astrocytes and microglia and were both spatially and temporally associated with intracellular cholesterol accumulation and neurodegeneration. Furthermore, levels of ubiquitinated proteins were higher in endosomal/lysosomal fractions of brains from Npc1(-/-) mice than from wild-type mice. Immunoblotting results showed that levels of LC3-II were significantly higher in brains of mutant than wild-type mice. Combined LC3 immunofluorescence and filipin staining showed that LC3 accumulated within filipin-labeled cholesterol clusters inside Purkinje cells. Electron microscopic examination revealed the existence of autophagic vacuole-like structures and multivesicles in brains from Npc1(-/-) mice. These results provide strong evidence that cholesterol accumulation-induced changes in autophagy-lysosome function are closely associated with neurodegeneration in NPC.
Figures
Figure 1
Cathepsin D levels in brain of Npc1+/+ and Npc1−/− mice during postnatal development. A: Representative images of blots of samples from 2-week-old animals labeled with anti-cathepsin D antibodies. Arrows correspond to the “single chain” isoform of cathepsin D, whereas lines indicate the “heavy form” of cathepsin D (see Results for details). B and C: Quantitative results for levels of single chain cathepsin D and heavy chain cathepsin D isoforms, respectively. Data are presented as percentage of values from Npc1+/+ mice and are means ± SEM. n = 5; *P < 0.05, and **P < 0.01.
Figure 2
Distribution of cathepsin D and B in cerebellum and thalamus of Npc1−/− mice during postnatal development. Cerebellar (A–D) and thalamic (E–L) tissue sections were prepared from Npc1+/+ (A, C, E, G, I, and K) and Npc1−/− (B, D, F, H, J, and L) mice at postnatal week 1 (A–F), 4 (K and L), and 8 (G–J) and were immunostained with anti-cathepsin D (A–J) or anti-cathepsin B (K and L) antibodies. Higher magnification images show that in Npc1+/+ mice (I) cathepsin D immunoreactive products are mainly located in small-sized granules, whereas in Npc1−/− mice they are present in larger puncta and their surrounding cytoplasmic structures (J). M shows immunoblots of cathepsin B-labeled samples from brainstem of 4-week-old mice. ml, molecular layer; pl, Purkinje layer; gl, granular layer; VPT, ventral posterior nucleus of thalamus. Scale bar = 50 μm (A and B); 12.5 μm (C–H and K and L).
Figure 3
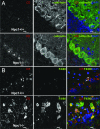
Cellular localization of cathepsin D in cerebellar cortex at 4 and 8 weeks postnatal. A: Double immunofluorescence staining using antibodies against cathepsin D (red) and calbindin (green) in cerebellum of 4-week-old Npc1+/+ (top panels) and Npc1−/− (bottom panels) mice. DAPI (blue) was included in the mounting medium to label nuclei. B: Double immunofluorescence staining using antibodies against cathepsin D (red) and F4/80 (green; a marker for microglia) in cerebellum of 8-week-old Npc1+/+ (top panels) and Npc1−/− (bottom panels) mice. *, Purkinje cells; arrows, Bergmann glia; mg, microglia. Scale bar = 10 μm.
Figure 4
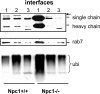
Abnormal protein distribution and ubiquitination in brain of Npc1−/− mice. Immunoblots of samples from different fractions labeled with anti-cathepsin D [single chain and heavy chain] and pro-cathepsin D (arrow in top panel), -rab7 (middle panel), or -ubiquitin (ubi, bottom panel) antibodies. Note the marked increase in cathepsin D in fraction 1 in samples from Npc1−/− mice. Note also the marked increase in levels of ubiquitinated proteins in endosomal/lysosomal fraction in the mutant mice. Interface 1 contains membrane from late endosome/lysosomes, interface 2 contains mainly early endosomes, and interface 3 contains other membrane structures. Western blots of subcellular fractions are representative of two experiments; each included four animals from each genotype. The results were very similar in both experiments.
Figure 5
LC3-II levels in brains of Npc1−/− mice during postnatal development. A: Representative images of blots labeled with anti-LC3 serum. Levels of LC3-II (B) and the ratio of LC3-II/LC3-I (C) are higher in mutant mice, especially in brainstem (BS) and cerebellum (CB); moderate increases were observed in cortex (CX) and hippocampus (Hipp). Data are presented as percentage of values from Npc1+/+ mice and are means ± SEM. n = 4; *P < 0.05, and **P < 0.01.
Figure 6
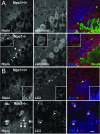
Cholesterol accumulation and sequestration of LC3 in Purkinje cells in Npc1−/− mice. A: Localization of filipin-stained free cholesterol (blue) in calbindin-immunopositive Purkinje cells (red) in cerebellum from 4-week-old Npc1+/+ (top) and Npc1−/− (bottom) mice. Note the accumulation of cholesterol in Purkinje cells in mutant mice but not in wild-type mice. Anti-NeuN (green) was used to label neurons. B: Combined LC3 immunostaining (red) with filipin staining (blue) in 8-week-old Npc1+/+ (top) and Npc1−/− (bottom) mice. Top panels show LC3-positive puncta present in Purkinje cells of 8-week-old Npc1+/+ mice; inset is a higher magnification image showing the subcellular distribution of LC3 puncta. Bottom panels show three-dimensional colocalization of LC3 with cholesterol in Purkinje cells of an 8-week-old mutant mouse. Scale bar = 10 μm.
Figure 7
Ultrastructure of Purkinje cells in Npc1+/+ mice. A: A Purkinje cell in the vicinity of granule cells in a 6-week-old Npc1+/+ mouse. B: Stacks of Golgi apparatus are present in a Purkinje cell. C and D: Lysosome-like structures exist in Purkinje cells. ER, endoplasmic reticulum; G, Golgi apparatus; L, lysosome; M, mitochondria; Ng, nucleus of granule cell; Npc, nucleus of Purkinje cell. Scale bars = 2 μm (A); 1 μm (B–D).
Figure 8
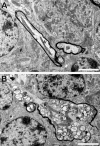
Ultrastructure of Purkinje cells in Npc1−/− mice. A: A Purkinje cell in a 6-week-old Npc1−/− mouse. Numerous vacuoles (arrowheads) of different sizes with various levels of electron-dense materials are present in the cytoplasm. B: Stacks of Golgi apparatus are clustered in the cytoplasm. C–H: Morphology of various membranous vacuoles. Some of them are with double membranes (arrowheads), whereas others have multilamellated structures (arrows). Scale bars = 2 μm (A); 1 μm (B–H).
Figure 9
Axonal pathology in Npc1−/− mice. A: A myelinated axon of a Purkinje cell axon exists in the vicinity of two granule cells in a 6-week-old wild-type mouse. B: An axonal spheroid in a myelinated Purkinje cell axon is located among granule cells in a 6-week-old Npc1−/− mouse. Note that a cluster of mitochondria is surrounded by vacuoles accumulated within the spheroid. Scale bars = 2 μm.
Figure 10
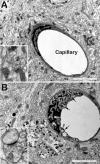
Capillary pathology in cerebellum of Npc1−/− mice. A capillary is surrounded by parallel fibers and synapses in the cerebellum of a 6-week-old wild-type (A) or a mutant (B) mouse. Note the membranous inclusions in the endothelial cell in the mutant mouse. Insets show synapses. Scale bars = 2 μm.
Similar articles
- Allopregnanolone treatment delays cholesterol accumulation and reduces autophagic/lysosomal dysfunction and inflammation in Npc1-/- mouse brain.
Liao G, Cheung S, Galeano J, Ji AX, Qin Q, Bi X. Liao G, et al. Brain Res. 2009 May 13;1270:140-51. doi: 10.1016/j.brainres.2009.03.027. Epub 2009 Mar 25. Brain Res. 2009. PMID: 19328188 Free PMC article. - Autophagic-lysosomal dysfunction and neurodegeneration in Niemann-Pick Type C mice: lipid starvation or indigestion?
Bi X, Liao G. Bi X, et al. Autophagy. 2007 Nov-Dec;3(6):646-8. doi: 10.4161/auto.5074. Autophagy. 2007. PMID: 17921694 - Increased activity and altered subcellular distribution of lysosomal enzymes determine neuronal vulnerability in Niemann-Pick type C1-deficient mice.
Amritraj A, Peake K, Kodam A, Salio C, Merighi A, Vance JE, Kar S. Amritraj A, et al. Am J Pathol. 2009 Dec;175(6):2540-56. doi: 10.2353/ajpath.2009.081096. Epub 2009 Nov 5. Am J Pathol. 2009. PMID: 19893049 Free PMC article. - Finding pathogenic commonalities between Niemann-Pick type C and other lysosomal storage disorders: Opportunities for shared therapeutic interventions.
Yañez MJ, Marín T, Balboa E, Klein AD, Alvarez AR, Zanlungo S. Yañez MJ, et al. Biochim Biophys Acta Mol Basis Dis. 2020 Oct 1;1866(10):165875. doi: 10.1016/j.bbadis.2020.165875. Epub 2020 Jun 6. Biochim Biophys Acta Mol Basis Dis. 2020. PMID: 32522631 Review. - Consequences of NPC1 and NPC2 loss of function in mammalian neurons.
Walkley SU, Suzuki K. Walkley SU, et al. Biochim Biophys Acta. 2004 Oct 11;1685(1-3):48-62. doi: 10.1016/j.bbalip.2004.08.011. Biochim Biophys Acta. 2004. PMID: 15465426 Review.
Cited by
- Viable neuronopathic Gaucher disease model in Medaka (Oryzias latipes) displays axonal accumulation of alpha-synuclein.
Uemura N, Koike M, Ansai S, Kinoshita M, Ishikawa-Fujiwara T, Matsui H, Naruse K, Sakamoto N, Uchiyama Y, Todo T, Takeda S, Yamakado H, Takahashi R. Uemura N, et al. PLoS Genet. 2015 Apr 2;11(4):e1005065. doi: 10.1371/journal.pgen.1005065. eCollection 2015 Apr. PLoS Genet. 2015. PMID: 25835295 Free PMC article. - Genistein Activates Transcription Factor EB and Corrects Niemann-Pick C Phenotype.
Argüello G, Balboa E, Tapia PJ, Castro J, Yañez MJ, Mattar P, Pulgar R, Zanlungo S. Argüello G, et al. Int J Mol Sci. 2021 Apr 19;22(8):4220. doi: 10.3390/ijms22084220. Int J Mol Sci. 2021. PMID: 33921734 Free PMC article. - Role of Endolysosomes in Skeletal Muscle Pathology Observed in a Cholesterol-Fed Rabbit Model of Alzheimer's Disease.
Chen X, Wagener JF, Ghribi O, Geiger JD. Chen X, et al. Front Aging Neurosci. 2016 Jun 8;8:129. doi: 10.3389/fnagi.2016.00129. eCollection 2016. Front Aging Neurosci. 2016. PMID: 27375475 Free PMC article. - Genetic dissection of a cell-autonomous neurodegenerative disorder: lessons learned from mouse models of Niemann-Pick disease type C.
Lopez ME, Scott MP. Lopez ME, et al. Dis Model Mech. 2013 Sep;6(5):1089-100. doi: 10.1242/dmm.012385. Epub 2013 Aug 1. Dis Model Mech. 2013. PMID: 23907005 Free PMC article. Review. - Altered Cholesterol Intracellular Trafficking and the Development of Pathological Hallmarks of Sporadic AD.
Chen X, Hui L, Soliman ML, Geiger JD. Chen X, et al. J Parkinsons Dis Alzheimers Dis. 2014;1(1):8. doi: 10.13188/2376-922x.1000002. J Parkinsons Dis Alzheimers Dis. 2014. PMID: 25621310 Free PMC article.
References
- Vance JE. Lipid imbalance in the neurological disorder, Niemann-Pick C disease. FEBS Lett. 2006;580:5518–5524. - PubMed
- Chang TY, Reid PC, Sugii S, Ohgami N, Cruz JC, Chang CC. Niemann-Pick type C disease and intracellular cholesterol trafficking. J Biol Chem. 2005;280:20917–20920. - PubMed
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, Nagle J, Polymeropoulos MH, Sturley SL, Ioannou YA, Higgins ME, Comly M, Cooney A, Brown A, Kaneski CR, Blanchette-Mackie EJ, Dwyer NK, Neufeld EB, Chang TY, Liscum L, Strauss JF, Ohno K, Zeigler M, Carmi R, Sokol J, Markie D, O’Neill RR, van Diggelen OP, Elleder M, Patterson MC, Brady RO, Vanier MT, Pentchev PG, Tagle DA. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. - PubMed
- Blom TS, Linder MD, Snow K, Pihko H, Hess MW, Jokitalo E, Veckman V, Syvanen AC, Ikonen E. Defective endocytic trafficking of NPC1 and NPC2 underlying infantile Niemann-Pick type C disease. Hum Mol Genet. 2003;12:257–272. - PubMed
- Garver WS, Heidenreich RA. The Niemann-Pick C proteins and trafficking of cholesterol through the late endosomal/lysosomal system. Curr Mol Med. 2002;2:485–505. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases