A protein interaction map of the mitotic spindle - PubMed (original) (raw)
. 2007 Oct;18(10):3800-9.
doi: 10.1091/mbc.e07-06-0536. Epub 2007 Jul 18.
Yuko Nakajima, Stefan Westermann, Ching Shang, Jung-Seog Kang, Crystal Goodner, Pantea Houshmand, Stanley Fields, Clarence S M Chan, David Drubin, Georjana Barnes, Tony Hazbun
Affiliations
- PMID: 17634282
- PMCID: PMC1995735
- DOI: 10.1091/mbc.e07-06-0536
A protein interaction map of the mitotic spindle
Jonathan Wong et al. Mol Biol Cell. 2007 Oct.
Abstract
The mitotic spindle consists of a complex network of proteins that segregates chromosomes in eukaryotes. To strengthen our understanding of the molecular composition, organization, and regulation of the mitotic spindle, we performed a system-wide two-hybrid screen on 94 proteins implicated in spindle function in Saccharomyces cerevisiae. We report 604 predominantly novel interactions that were detected in multiple screens, involving 303 distinct prey proteins. We uncovered a pattern of extensive interactions between spindle proteins reflecting the intricate organization of the spindle. Furthermore, we observed novel connections between kinetochore complexes and chromatin-modifying proteins and used phosphorylation site mutants of NDC80/TID3 to gain insights into possible phospho-regulation mechanisms. We also present analyses of She1p, a novel spindle protein that interacts with the Dam1 kinetochore/spindle complex. The wealth of protein interactions presented here highlights the extent to which mitotic spindle protein functions and regulation are integrated with each other and with other cellular activities.
Figures
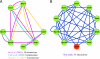
Figure 1.
Comparison of intra-Dam1 complex interactions detected from genome-wide and focused two-hybrid screens. Protein interaction networks of subunits within the Dam1 complex derived from yeast two-hybrid studies and an in vitro expression experiment were generated with Cytoscape network visualization software. The bait construct for Ask1p (shown in red) was lethal to yeast and could not be screened. (A) The interactions reported from previous comprehensive two-hybrid screens identified seven of the 10 Dam1 complex subunits. (B) The network of interactions found by this study identified all 10 subunits of the Dam1 complex. The very high number of interactions detected between subunits is consistent with their association as a protein complex.
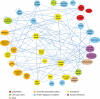
Figure 2.
A simplified spindle protein interaction network. This simplified network includes proteins with demonstrated spindle or chromosome functions as well as uncharacterized proteins that interact with multiple spindle proteins. Proteins that belong to the same complex or functional process are grouped into single nodes. cmplx, complex; APC, anaphase-promoting complex; CK1, casein kinase I; CKII, casein kinase II; CAF-I, chromatin assembly factor I; CRC, chromatin remodeling complex; CDK, cyclin-dependent kinase; HATs, histone acetyltransferases; HDACs, histone deacetylases; MAPs, microtubule-associated proteins; PP1, protein phosphatase I; PP2A, protein phosphatase 2A.
Figure 3.
She1-3GFP localizes to the mitotic spindle and the bud neck. (A) Localization of She1-3GFP during metaphase and anaphase. (B) She1-3GFP (green) colocalizes with Duo1-RFP (red) on the mitotic spindle. (C) She1-3GFP (green) colocalizes with Sli15-RFP (red) on microtubules. Bar, 4 μm.
Figure 4.
Comparison of protein interaction maps of Ndc80p phospho-mutants. The protein interaction maps shown here summarize the results of yeast two-hybrid screens performed using wild-type Ndc80p and phospho-mutants that mimic the phosphorylation and dephosphorylation of the four N-terminal Ipl1/Aurora B consensus sites, as baits. The color of the nodes corresponds with their GO Process classification. The interactions are classified as being common to all three alleles (blue lines), exclusive of the ndc80-4A allele (green lines), or exclusive of the ndc80-4D allele (orange lines). The protein interaction maps shown here were generated with OSPREY (
http://biodata.mshri.on.ca/osprey
; Breitkreutz et al., 2003).
Similar articles
- Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p.
Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. Shang C, et al. Mol Biol Cell. 2003 Aug;14(8):3342-55. doi: 10.1091/mbc.e02-11-0765. Epub 2003 May 3. Mol Biol Cell. 2003. PMID: 12925767 Free PMC article. - Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity.
Enquist-Newman M, Cheeseman IM, Van Goor D, Drubin DG, Meluh PB, Barnes G. Enquist-Newman M, et al. Mol Biol Cell. 2001 Sep;12(9):2601-13. doi: 10.1091/mbc.12.9.2601. Mol Biol Cell. 2001. PMID: 11553702 Free PMC article. - Molecular architecture and assembly of the yeast kinetochore MIND complex.
Maskell DP, Hu XW, Singleton MR. Maskell DP, et al. J Cell Biol. 2010 Sep 6;190(5):823-34. doi: 10.1083/jcb.201002059. J Cell Biol. 2010. PMID: 20819936 Free PMC article. - Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans.
Westermann S, Schleiffer A. Westermann S, et al. Trends Cell Biol. 2013 Jun;23(6):260-9. doi: 10.1016/j.tcb.2013.01.010. Epub 2013 Mar 5. Trends Cell Biol. 2013. PMID: 23481674 Review. - Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1.
Scarfone I, Piatti S. Scarfone I, et al. Small GTPases. 2015 Oct 2;6(4):196-201. doi: 10.1080/21541248.2015.1109023. Small GTPases. 2015. PMID: 26507466 Free PMC article. Review.
Cited by
- Constitutive dynein activity in She1 mutants reveals differences in microtubule attachment at the yeast spindle pole body.
Bergman ZJ, Xia X, Amaro IA, Huffaker TC. Bergman ZJ, et al. Mol Biol Cell. 2012 Jun;23(12):2319-26. doi: 10.1091/mbc.E12-03-0223. Epub 2012 Apr 25. Mol Biol Cell. 2012. PMID: 22535527 Free PMC article. - Slk19 clusters kinetochores and facilitates chromosome bipolar attachment.
Richmond D, Rizkallah R, Liang F, Hurt MM, Wang Y. Richmond D, et al. Mol Biol Cell. 2013 Mar;24(5):566-77. doi: 10.1091/mbc.E12-07-0552. Epub 2013 Jan 2. Mol Biol Cell. 2013. PMID: 23283988 Free PMC article. - Hec1 contributes to mitotic centrosomal microtubule growth for proper spindle assembly through interaction with Hice1.
Wu G, Wei R, Cheng E, Ngo B, Lee WH. Wu G, et al. Mol Biol Cell. 2009 Nov;20(22):4686-95. doi: 10.1091/mbc.e08-11-1123. Epub 2009 Sep 23. Mol Biol Cell. 2009. PMID: 19776357 Free PMC article. - Nuclear transporters in a multinucleated organism: functional and localization analyses in Aspergillus nidulans.
Markina-Iñarrairaegui A, Etxebeste O, Herrero-García E, Araújo-Bazán L, Fernández-Martínez J, Flores JA, Osmani SA, Espeso EA. Markina-Iñarrairaegui A, et al. Mol Biol Cell. 2011 Oct;22(20):3874-86. doi: 10.1091/mbc.E11-03-0262. Epub 2011 Aug 31. Mol Biol Cell. 2011. PMID: 21880896 Free PMC article. - The microtubule-associated protein She1 coordinates directional spindle positioning by spatially restricting dynein activity.
Ecklund KH, Bailey ME, Kossen KA, Dietvorst CK, Asbury CL, Markus SM. Ecklund KH, et al. J Cell Sci. 2021 Dec 1;134(23):jcs258510. doi: 10.1242/jcs.258510. Epub 2021 Dec 2. J Cell Sci. 2021. PMID: 34854468 Free PMC article.
References
- Ayscough K. R., Drubin D. G. Cell Biology: A Laboratory Handbook. Vol. 2. San Diego: Academic Press; 1998. Immunofluorescence microscopy of yeast cells; pp. 477–485.
- Biddick R., Young E. T. Yeast mediator and its role in transcriptional regulation. CR Biol. 2005;328:773–782. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases