Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2 - PubMed (original) (raw)
Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2
Zheng Sun et al. Mol Cell Biol. 2007 Sep.
Abstract
The transcription factor Nrf2 regulates cellular redox homeostasis. Under basal conditions, Keap1 recruits Nrf2 into the Cul3-containing E3 ubiquitin ligase complex for ubiquitin conjugation and subsequent proteasomal degradation. Oxidative stress triggers activation of Nrf2 through inhibition of E3 ubiquitin ligase activity, resulting in increased levels of Nrf2 and transcriptional activation of Nrf2-dependent genes. In this study, we identify Keap1 as a key postinduction repressor of Nrf2 and demonstrate that a nuclear export sequence (NES) in Keap1 is required for termination of Nrf2-antioxidant response element (ARE) signaling by escorting nuclear export of Nrf2. We provide evidence that ubiquitination of Nrf2 is carried out in the cytosol. Furthermore, we show that Keap1 nuclear translocation is independent of Nrf2 and the Nrf2-Keap1 complex does not bind the ARE. Collectively, our results suggest the following mechanism of postinduction repression: upon recovery of cellular redox homeostasis, Keap1 translocates into the nucleus to dissociate Nrf2 from the ARE. The Nrf2-Keap1 complex is then transported out of the nucleus by the NES in Keap1. Once in the cytoplasm, the Keap1-Nrf2 complex associates with the E3 ubiquitin ligase, resulting in degradation of Nrf2 and termination of the Nrf2 signaling pathway. Hence, postinduction repression of the Nrf2-mediated antioxidant response is controlled by the nuclear export function of Keap1 in alliance with the cytoplasmic ubiquitination and degradation machinery.
Figures
FIG. 1.
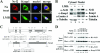
The cytoplasmic-nuclear trafficking of Nrf2 and Keap1. (A) MDA-MB-231 cells were treated with 10 nM of LMB for 3 h. Subcellular localization of Nrf2 and Keap1 was detected by indirect immunofluorescence staining with anti-Nrf2 and anti-Keap1 antibodies. (B) Nuclear and cytoplasmic fractions of mock- or LMB-treated MDA-MB-231 cells were subjected to immunoblot analysis with anti-Nrf2 and anti-Keap1 antibodies (α-Nrf2 and α-Keap1) α-Lamin A, anti-lamin A antibody; α-tubulin, antitubulin antibody. (C) Upper panel, six discrete domains of the Nrf2 proteins are designated Neh2, Neh4, Neh5, Neh6, Neh1, and Neh3. The two putative NESs in Nrf2 are located in the Neh5 and Neh1 domains. The amino acid sequences of the two NESs are indicated in the single-letter code below the Nrf2 drawing. The residues relevant to the present work are in boldface, and the mutations introduced into the Nrf2 protein are indicated. Lower panel, Keap1 contains five domains: N-terminal, BTB, Linker, Kelch, and C-terminal. The NES in Keap1 is in the Linker domain. The NES sequence with a cluster of hydrophobic residues is shown, and the four hydrophobic residues are replaced by alanines. (D) COS1 cells were cotransfected with expression vectors for the indicated Nrf2 and Keap1 proteins. Total lysates were analyzed by immunoblotting with anti-HA and anti-CBD antibodies (α-HA and α-CBD) for detection of Nrf2 and Keap1 proteins (upper two panels). The lysates were incubated with chitin beads. Following washing, protein complexes bound to chitin beads were eluted by heating in sample buffer and subjected to immunoblot analysis with anti-HA and anti-CBD antibodies (lower two panels).
FIG. 2.
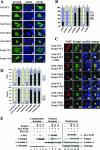
The NES in Keap1 controls nuclear export of Nrf2. (A) NIH 3T3 cells were singly transfected with an expression vector for the indicated Nrf2 or Keap1 protein. Subcellular localization of Nrf2 or Keap1 was determined by indirect immunofluorescence analysis using anti-HA for Nrf2 (panels A, D, G, J, and M) or anti-CBD for Keap1 (panels P and S). The nucleus was visualized by Hoechst staining (panels B, E, H, K, N, Q, and T). (B) Subcellular distribution of the Nrf2 or Keap1 protein in singly transfected cells (the representative image is shown in A) was determined by examining at least 100 positive cells. Percentages of cells that localized predominantly in the cytosol (C), the whole cell (W), or the nucleus (N) were presented as a bar graph. (C) NIH 3T3 cells were cotransfected with expression vectors for the indicated Nrf2 and Keap1 proteins. Subcellular localization of the Nrf2 and Keap1 proteins was determined by double-labeling indirect immunofluorescence assay using anti-HA (panels 1, 5, 9, 13, 17, 21, and 25) and anti-CBD antibodies (panels 2, 6, 10, 14, 18, 22, and 26). The nucleus was visualized by Hoechst staining (panels 3, 7, 11, 15, 19, 23, and 27). Colocalization of the Nrf2 and Keap1 proteins is indicated by the presence of yellow in the merged images (panels 4, 8, 12, 16, 20, 24, and 28). (D) Subcellular localization of the Nrf2 and Keap1 proteins in double-transfected cells (the same slides were used for C) were examined and counted in the same way as in B except that the data were collected from 100 cells that are positive for both Nrf2 and Keap1 proteins. (E) Nuclear and cytoplasmic proteins were isolated from MDA-231 cells cotransfected with expression vectors for Nrf2 and either Keap1-WT or Keap1-NES. The transfected cells were either left untreated or treated with 10 μM MG132 for 4 h prior to subcellular fractionation. Nuclear and cytoplasmic proteins derived from equal numbers of cells were electrophoresed through a 7.5% SDS-polyacrylamide gel and subjected to immunoblot analysis using anti-HA, anti-Keap1, antitubulin, or anti-lamin A antibodies. α, anti.
FIG. 3.
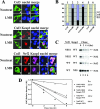
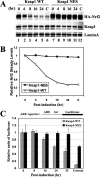
Ubiquitination of Nrf2, mediated by the Keap1-Cul3-Rbx1 E3 ubiquitin ligase, occurs in the cytosol. (A) NIH 3T3 cells were singly transfected with an expression vector for Flag-Cul3 (upper panel), cotransected with expression vectors for both Flag-Cul3 and Keap1 (middle panel), or cotransfected with expression vectors for Flag-Cul3, Keap1, and HA-Nrf2 (lower panel). Subcellular localization of Cul3, Nrf2, or Keap1 was determined by indirect immunofluorescence analysis using anti-Flag for Cul3 (panels A, D, G, K, O, and T), anti-HA for Nrf2 (panels P and U), or anti-Keap1 for Keap1 (panels H, L, Q, and V). Nontreat, nontreated; LMB, LMB treatment. (B) Subcellular distribution of Cul3, Nrf2, and Keap1 in singly transfected cells in the absence or presence of LMB was determined by indirect immunofluorescence staining. At least 100 positive cells were examined. Percentages of cells that localized predominantly in the cytosol (C), the whole cell (W), or the nucleus (N) were presented as a bar graph. K-WT, Keap1-WT; N-WT, Nrf2-WT; +LMB, LMB treatment. (C) MDA-MB-231 cells were cotransfected with expression vectors for the indicted Nrf2 and Keap1 proteins. Fifty micrograms per milliliter cycloheximide was added 36 h after transfection. Total cell lysates were collected at the indicated time points following cycloheximide treatment and subjected to immunoblot analysis with anti-HA antibodies. (D) The relative intensities of the Nrf2 bands were quantified by the ChemiDoc XRS gel documentation system from Bio-Rad and plotted on a semilog scale. The amount of Nrf2 present at the beginning of cycloheximide treatment was set at 1. The half-life of Nrf2 in each group was indicated.
FIG. 4.
Keap1-mediated nuclear export of Nrf2 is required for ubiquitination of Nrf2. (A) In vivo ubiquitination of Nrf2 in the presence of Keap1-WT or Keap1-NES was measured in MDA-MB-231 cells cotransfected with expression vectors for HA-ubiquitin, Nrf2, and the indicated Keap1 protein. The transfected cells were left untreated or treated with tBHQ for 4 h. Cells were lysed under denaturing conditions, and small aliquots of total lysates were used for immunoblot analysis with anti-Nrf2 (α-Nrf2), anti-CBD (α-CBD), and antitubulin (α-Tub) antibodies (lower panel). Anti-Nrf2 immunoprecipitates (IP) were analyzed by immunoblotting with anti-HA antibodies for detection of ubiquitinated Nrf2 (upper panel). (B) In vitro ubiquitination of Nrf2 in the presence of Keap1-WT and Keap1-NES was measured in COS-1 cells transfected with expression vectors for Nrf2, HA-Cul3, Myc-Rbx1, and the indicated Keap1 protein. Lysates from one 100-mm dish were incubated with chitin beads, and the bound proteins were eluted in sample buffer by boiling and subjected to immunoblot analysis with anti-HA, anti-CBD, and anti-Myc antibodies (α-HA, α-CBD, and α-Myc) (lower panel). Chitin bead-bound proteins from another 100-mm dish were incubated with purified E1, E2-UbcH5a, ubiquitin, and ATP. The chitin beads were pelleted and washed. Proteins that were eluted from the beads after boiling were immunoprecipitated with anti-Nrf2 antibodies (α-Nrf2), and Nrf2 immunoprecipitates were analyzed by immunoblotting with antiubiquitin antibodies (α-Ub) for detection of ubiquitin-conjugated Nrf2 (upper panel).
FIG. 5.
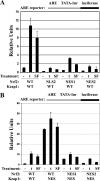
Nuclear export of Nrf2 is required for repression of Nrf2-dependent transcriptional activity. (A and B) MDA-MB-231 cells were cotransfected with plasmids containing an ARE-dependent firefly luciferase reporter gene and expression plasmids for the indicated Nrf2 and Keap1 proteins. A plasmid encoding Renilla luciferase driven by the herpes simplex virus thymidine kinase promoter was included in all transfections to normalize transfection efficiency. The transfected cells were exposed to dimethyl sulfoxide (−), 50 μM tBHQ (t), or 10 μM SF (SF) for 16 h prior to analysis of firefly and Renilla luciferase activities in cell lysates. All samples were duplicated for each experiment, and the data shown represent the means for three independent experiments. The error bars indicate the standard deviations for the three experiments. Please note the difference in the scale of relative units between panels A and B.
FIG. 6.
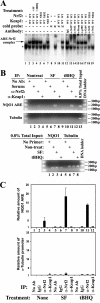
Nrf2, not Keap1, associates with ARE. (A) In vitro interaction of Nrf2 or Keap1 with the ARE was analyzed by EMSA. MDA-MB-231 cells cotransfected with either an empty vector or expression vectors for the indicated Nrf2 and Keap1 proteins were either left untreated, treated with 10 μM SF, or treated with 100 μM tBHQ. Nuclear fractions were extracted and incubated with a 32P-labeled ARE-containing oligonucleotide in the absence or presence of either the unlabeled competing oligonucleotides or antibodies. The protein-DNA complexes were size separated on a nondenaturing polyacrylamide gel. The arrow indicates the position of the ARE-Nrf2 complexes. The asterisk indicates an ARE binding complex that does not contain Nrf2. Two different types of Nrf2 antibodies were used. The one labeled with an asterisk has a higher concentration. α-Nrf2, α-Keap, α-p300, and α-Gal4, anti-Nrf2, -Keap1, -p300, and -Gal4 antibodies. (B and C) In vivo interaction of Nrf2 or Keap1 with the ARE was determined by a ChIP assay. MDA-MB-231 cells were left untreated, treated with tBHQ, or treated with SF. DNA-protein complexes were cross-linked and immunoprecipitated with the indicated antibodies. Amounts of DNA containing the NQO1-ARE or the tubulin promoter were semiquantified by real-time PCR amplification with a primer pair flanking the human NQO1 ARE sequence (upper panel) or a primer pair specific for the human tubulin promoter (middle panel). No Ab, no antibody. A 0.8% proportion of total input DNA for immunoprecipitation was included as positive controls for real-time PCR amplification (lower panel). (C) Amounts of immunoprecipitated NQO1 ARE (upper panel) and the tubulin promoter (lower panel) were semiquantified by real-time PCR amplification and presented as a bar graph using the LightCycler 480 software.
FIG. 7.
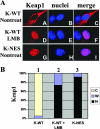
Nuclear import of Keap1 is independent of Nrf2. (A) Nrf2−/− mouse embryonic fibroblast cells were singly transfected with an expression vector for either Keap1-WT (panels A to F) or Keap1-NES (panels G to I). Cells were left untreated (Nontreat) or treated with LMB (panels D to F) for 3 h. Subcellular localization of the indicated Keap1 protein was determined by an indirect immunofluorescence analysis using anti-CBD antibodies. K-WT, Keap1-WT. (B) The same slides from panel A were used to count at least 100 positive cells. Percentages of cells that localized predominantly in the cytosol (C), the whole cell (W), or the nucleus (N) were presented as a bar graph.
FIG. 8.
Keap1 confers postinduction repression of Nrf2 by escorting nuclear export of Nrf2. (A and B) Postinduction repression of the steady-state levels of Nrf2 was accessed in MDA-MB-231 cells cotransfected with an expression vector for Nrf2 and an expression vector for either Keap1-WT or Keap1-NES. Cells were treated with 100 μM tBHQ for 4 h. After removal of tBHQ by washing, cells were further incubated in normal medium for the indicated time periods. Total cell lysates were subjected to immunoblot analysis with anti-HA, anti-CBD, and anti-lamin A antibodies. (B) The relative intensities of the Nrf2 bands were quantified by the ChemiDoc XRS gel documentation system from Bio-Rad and plotted on a linear graph. (C) Postinduction repression of the Nrf2-dependent transcriptional activity was determined in MDA-MB-231 cells cotransfected with plasmids encoding an ARE-firefly luciferase, thymidine kinase-Renilla luciferase, Nrf2, and the indicated Keap1 protein. The transfected cells were exposed to 50 μM tBHQ for 16 h. Following removal of tBHQ, cells were further incubated in normal medium for the indicated time periods prior to measurement of firefly and Renilla luciferase activities. The experiment was repeated three times, and standard deviations are shown as error bars. Untreat, untreated.
FIG. 9.
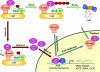
Schematic model of Nrf2 regulation by Keap1. Keap1 is a key regulator of the Nrf2 signaling pathway and serves as a molecular switch to turn on and off the Nrf2-mediated antioxidant response. (i) The switch is in off position: under basal conditions, Keap1, functioning as an E3 ubiquitin ligase, constantly targets Nrf2 for ubiquitination and degradation. As a consequence, there are minimal levels of Nrf2. (ii) The switch is turned on: oxidative stress or chemopreventive compounds inhibit activity of the Keap1-Cul3-Rbx1 E3 ubiquitin ligase, resulting in increased levels of Nrf2 and activation of its downstream target genes. (iii) The switch is turned off again: Upon recovery of cellular redox homeostasis, Keap1 travels into the nucleus to remove Nrf2 from the ARE. The Nrf2-Keap1 complex is then transported out of the nucleus by the NES in Keap1. In the cytosol, the Nrf2-Keap1 complex associates with the Cul3-Rbx1 core ubiquitin machinery, leading to degradation of Nrf2. For clarity, the constitutive cytoplasmic-nuclear shuttling of Nrf2, Keap1, and the complex is omitted.
Similar articles
- Review of molecular mechanisms involved in the activation of the Nrf2-ARE signaling pathway by chemopreventive agents.
Giudice A, Arra C, Turco MC. Giudice A, et al. Methods Mol Biol. 2010;647:37-74. doi: 10.1007/978-1-60761-738-9_3. Methods Mol Biol. 2010. PMID: 20694660 Review. - Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism.
Velichkova M, Hasson T. Velichkova M, et al. Mol Cell Biol. 2005 Jun;25(11):4501-13. doi: 10.1128/MCB.25.11.4501-4513.2005. Mol Cell Biol. 2005. PMID: 15899855 Free PMC article. - Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex.
Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Zhang DD, et al. Mol Cell Biol. 2004 Dec;24(24):10941-53. doi: 10.1128/MCB.24.24.10941-10953.2004. Mol Cell Biol. 2004. PMID: 15572695 Free PMC article. - The Keap1-Nrf2-antioxidant response element pathway: a review of its regulation by melatonin and the proteasome.
Vriend J, Reiter RJ. Vriend J, et al. Mol Cell Endocrinol. 2015 Feb 5;401:213-20. doi: 10.1016/j.mce.2014.12.013. Epub 2014 Dec 17. Mol Cell Endocrinol. 2015. PMID: 25528518 Review.
Cited by
- Dietary Oxidative Distress: A Review of Nutritional Challenges as Models for Poultry, Swine and Fish.
Bacou E, Walk C, Rider S, Litta G, Perez-Calvo E. Bacou E, et al. Antioxidants (Basel). 2021 Mar 27;10(4):525. doi: 10.3390/antiox10040525. Antioxidants (Basel). 2021. PMID: 33801670 Free PMC article. Review. - The Nrf2-ARE pathway: a valuable therapeutic target for the treatment of neurodegenerative diseases.
Joshi G, Johnson JA. Joshi G, et al. Recent Pat CNS Drug Discov. 2012 Dec;7(3):218-29. doi: 10.2174/157488912803252023. Recent Pat CNS Drug Discov. 2012. PMID: 22742419 Free PMC article. Review. - Oxidant-induced cell death and Nrf2-dependent antioxidative response are controlled by Fra-1/AP-1.
Vaz M, Machireddy N, Irving A, Potteti HR, Chevalier K, Kalvakolanu D, Reddy SP. Vaz M, et al. Mol Cell Biol. 2012 May;32(9):1694-709. doi: 10.1128/MCB.06390-11. Epub 2012 Mar 5. Mol Cell Biol. 2012. PMID: 22393254 Free PMC article. - A systems biology approach to defining regulatory mechanisms for cartilage and tendon cell phenotypes.
Mueller AJ, Tew SR, Vasieva O, Clegg PD, Canty-Laird EG. Mueller AJ, et al. Sci Rep. 2016 Sep 27;6:33956. doi: 10.1038/srep33956. Sci Rep. 2016. PMID: 27670352 Free PMC article. - Non-thermal plasma activates human keratinocytes by stimulation of antioxidant and phase II pathways.
Schmidt A, Dietrich S, Steuer A, Weltmann KD, von Woedtke T, Masur K, Wende K. Schmidt A, et al. J Biol Chem. 2015 Mar 13;290(11):6731-50. doi: 10.1074/jbc.M114.603555. Epub 2015 Jan 14. J Biol Chem. 2015. PMID: 25589789 Free PMC article.
References
- Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Cap′n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. - PubMed
- Aoki, Y., H. Sato, N. Nishimura, S. Takahashi, K. Itoh, and M. Yamamoto. 2001. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol. Appl. Pharmacol. 173:154-160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical