An essential switch in subunit composition of a chromatin remodeling complex during neural development - PubMed (original) (raw)
An essential switch in subunit composition of a chromatin remodeling complex during neural development
Julie Lessard et al. Neuron. 2007.
Abstract
Mammalian neural stem cells (NSCs) have the capacity to both self-renew and to generate all the neuronal and glial cell-types of the adult nervous system. Global chromatin changes accompany the transition from proliferating NSCs to committed neuronal lineages, but the mechanisms involved have been unclear. Using a proteomics approach, we show that a switch in subunit composition of neural, ATP-dependent SWI/SNF-like chromatin remodeling complexes accompanies this developmental transition. Proliferating neural stem and progenitor cells express complexes in which BAF45a, a Krüppel/PHD domain protein and the actin-related protein BAF53a are quantitatively associated with the SWI2/SNF2-like ATPases, Brg and Brm. As neural progenitors exit the cell cycle, these subunits are replaced by the homologous BAF45b, BAF45c, and BAF53b. BAF45a/53a subunits are necessary and sufficient for neural progenitor proliferation. Preventing the subunit switch impairs neuronal differentiation, indicating that this molecular event is essential for the transition from neural stem/progenitors to postmitotic neurons. More broadly, these studies suggest that SWI/SNF-like complexes in vertebrates achieve biological specificity by combinatorial assembly of their subunits.
Figures
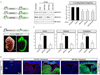
Figure 1.. BAF45 family proteins are subunits of neural SWI/SNF-like BAF complexes
A. Immunoprecipitation and Western blot analysis of Brg/Brm proteins in P0 mouse nuclear extracts using anti-Brg/Brm affinity purified antibodies. kDa, kilodaltons. B. Strategy for purification and sequencing of neural SWI/SNF-like BAF complexes. Highly stringent conditions were used to isolate only the core subunits of the complexes plus tightly associated proteins (see supplemental experimental procedures). ESI-MS/MS, Electrospray Ionization- Tandem Mass Spectrometry. C. Silver-stain analysis of immunopurified (anti-Brg/Brm) endogenous neural BAF complexes in P0 mouse brain nuclear extracts. D. Peptides identified from the BAF45 and BAF53 families of subunits by mass spectrometry. The numbers in parentheses are the numbers of peptides corresponding to unique protein entries. MW, calculated molecular weight; aa, amino acids. E. Conservation and divergence among mouse BAF45 family members. Conserved amino acids are highlighted in blue. The C2H2-type Krüppel-like domains are boxed in red and the PHD domains of the d4 domain are boxed in green. Red and blue stars indicate the conserved cysteine and histidine residues, respectively. The putative nuclear localization sequence is underlined in blue and is displaced 28 aa C-terminal in BAF45a. Note that only the first C3H motif of the PHD1 domain is conserved in BAF45c. F. The C2H2-type Krüppel-like zinc finger motif is not conserved in BAF45a. The conserved cysteine and histidine residues are labeled with red and blue stars, respectively. Note their absence in BAF45a protein.
Figure 2.. The subunit composition of neural BAF complexes changes as cells develop from neural progenitors to post-mitotic neurons
A. Western blot analyses of immunopurified (anti-Brg/Brm) neural BAF complexes in P0 mouse brain nuclear extracts. B. BAF45a, BAF45b and BAF45c are dedicated to neural BAF complexes. Serial immunodepletions were performed on anti-Brg/Brm columns in P0 brain nuclear extracts. Supernatants were blotted with anti-BAF antibodies. Hsp90, heat shock protein 90; id, immunodepletion. C. BAF45a, BAF45b and BAF45c co-sediment with other components of neural BAF complexes. E15.5 brain nuclear extracts were separated by centrifugation on glycerol gradients (10 to 30 %) and isolated fractions analyzed by Western blotting. D. Sequential expression of BAF45a, BAF53a and BAF45b, BAF45c and BAF53b during neurogenesis. Brain nuclear extracts were isolated at different developmental stages and blotted with BAF45, BAF53 and Brg specific antibodies. E. BAF45a and BAF53a are expressed specifically in neural progenitors and are replaced by the BAF45b/c and BAF53b homologous subunits in post-mitotic neurons. Immunostaining on cross sections of E12.5 spinal cords together with progenitor-(LeX) and neuronal-specific (TuJ1 and NeuN) markers. TuJ1, antibody against β-tubulin type III; DAPI, 4′,6-Diamidino-2-phenylindole.
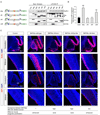
Figure 3.. BAF45a and BAF53a are necessary and sufficient for the proliferation of neural progenitors
A. shRNA-mediated knockdown of BAF45a and/or BAF53a impairs the proliferative activity of neural progenitors in E14.5 adherent cortical cultures. Western blot analyses of protein extracts from these cultures (3 div) are shown. Hsp90 is a loading control. E14.5 cortical cells electroporated (Amaxa Inc, MD) with shRNA or control vectors were cultured in serum-containing media (4 div), stained with anti-LeX or anti-nestin and BrdU antibodies and analyzed by flow cytometry. BrdU was added 2 hrs before analysis. The percentage of dividing GFP+ BrdU+ neural progenitors in these cultures (n=3 independent experiments) is shown. Results are expressed as mean ±SD. **, p<0.01. See supplemental information for shRNA sequences. CMV, cytomegalovirus immediate early enhancer/promoter. B. _Nestin_-driven _BAF45a_ IRES-_GFP_ expression in mouse E13.5 transient transgenics. Nestin, nestin-gene regulatory elements; IRES, internal ribosomal entry site. Note that the BAF45a and BAF45b expression patterns and levels were similar in all the _BAF45a_ and _BAF45b_ transient transgenic mice analyzed. C. Quantification of the number of proliferating (H3P+) neural progenitors in the cortex, midbrain and cerebellum of _BAF45a_ and _BAF45b_ transient transgenic mice (n=5 and n=3 embryos, respectively) relative to control littermates. H3P+ cells (red) were manually counted using digitalized pictures of comparative brain sagittal sections stained with anti-H3P antibody (n>6 pictures per embryo). Results are expressed as mean ± SD. **, p<0.01. H3P, anti-phosphorylated Histone 3 (Ser10). D. Increased number of proliferating (H3P+) neural progenitors in the developing cortices and basal ganglia of E13.5 BAF45a transgenic embryos relative to BAF45b transgenic and control embryos. Comparable sagittal sections were stained with anti-H3P antibodies (red) and DAPI (blue). Representative images are shown. BG, basal ganglia; CT, cortex; V, ventricle.
Figure 4.. The N-terminus and Krüppel-like domains of BAF45a are essential to induce neural progenitor proliferation
A. The N-terminal, Krüppel-like and d4 domains of BAF45a are not required for its assembly within npBAF complexes in P19 cells. Schematic representations of the transgenes are shown. Nuclear extracts of transfected P19 cells expressing similar levels of the FLAG-tagged proteins (β-actin promoter) were immunoprecipitated with M2 (anti-FLAG) agarose beads and analyzed by Western blotting. Note the lower detection level of the BAF45a ΔKrüppel-like protein (deletion aa 204–246) with the α-BAF45a antibody (raised against aa 180 to 282). *; non-specific band recognized by the FLAG antibody. B. Number of M-phase (H3P+) cerebellar progenitors in E14.5 Nestin-driven BAF45a (n=3), BAF45a ΔN-terminal (n=2), BAF45a ΔKrüppel-like (n=3) and BAF45a Δd4 domain (n=3) transient transgenic mice relative to control littermates. H3P+ cells (red) were manually counted using digitalized pictures of comparative brain sagittal sections stained with anti-H3P antibody (n>6 pictures per embryo). Results are expressed as mean ± SD. **, p<0.01 C. Dividing (BrdU+ and H3P+) neural progenitors in the developing cerebellums and midbrains of _Nestin_-driven BAF45a and BAF45a deletion mutant transgenic mice (E14.5, 1 hr BrdU pulse). Comparable sagittal sections were stained with anti-BrdU (red) or anti-H3P (red) antibodies and DAPI (blue). Number of independent transgenic lines: BAF45a wild-type (n= 3), BAF45a ΔN-terminal (n= 2), BAF45a ΔKrüppel-like (n= 3) and BAF45a Δd4 domain (n=3). Representative images are shown. EGL, external granular layer; M, midbrain.
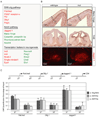
Figure 5.. Brg is essential for the self-renewal and maintenance of neural stem/progenitor cells
A. Brg deletion does not affect the expression of Brm and other npBAF subunits. Strategy for deletion of Brg in neural progenitors. Genomic structure of the floxed Brg allele from exon 15 through 18 encoding part of the conserved ATPase domain (Sumi-Ichinose et al., 1997). Two LoxP sites (empty arrowheads) were inserted in introns 15 and 17. The Cre recombinase expressed in neural progenitors by the Nestin-Cre transgene recombines the two LoxP sites and results in the deletion of exons 16 and 17. If exon 15 splices to exon 18, translation of the Brg protein stops at aa 819. Western blot analysis of BAF subunits in cellular extracts from E13.5 control and Brg mutant cortices. Hsp90 is a loading control. B. Brg is deleted in neural stem/progenitor cells by E12.5. Sagittal sections of E12.5 control (wt) and Brg mutant frontal cortices were immunostained with specific anti-Brg antibodies (red). mut, mutant. C. Brg mutant brains are smaller and lack a cerebellum (arrows). Hematoxylin and eosin (H&E) staining of sagittal sections of E18.5 wt and Brg mutant brains. D. Progressive depletion of cortical neural progenitors in the Brgf/f;Nestin-Cre mice during embryogenesis. Upper panels: comparable sections of E12.5 and E17.5 cortices stained with anti-Nestin (red), anti-H3P (green) antibodies and DAPI (blue). Note the near absence of Nestin+ cells in E17.5 Brg mutant cortices. Lower panels: similar cortical regions stained with TuJ1 antibody (red). E. Defective neurosphere formation by _Brg_-deficient cortical cells. Primary neurospheres were cultured from dissociated E13.5 control and mutant cortices for 7 div. Secondary spheres were cultured from dissociated primary spheres (2 wt neurospheres and 20 Brg mutant neurospheres) for another 4 div. F. _Brg_-deficient progenitors proliferate slower than wild-type progenitors. Brg mutant and control E14.5 cortical cells were cultured in serum-containing media for 15 hrs (1 div), stained with anti-LeX and BrdU antibodies and analyzed by flow cytometry. BrdU was added 3 hrs before the analysis. Representative images of n=3 independent experiments performed in duplicate are shown. G. Brg is not required for glia proliferation in vitro. Brg mutant and control E16.5 cortical cells were cultured in serum-containing media (3 div) with or without CNTF (100 ng/ml), stained with anti-GFAP and BrdU antibodies and analyzed by flow cytometry. Results are expressed as mean ± SD. Western blot analyses of Brg (H-88) and GFAP protein levels in these cultures are shown. CNTF, ciliary neurotropic factor; GFAP, glial fibrillary acidic protein.
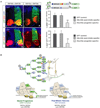
Figure 6.. Brg-containing npBAF complexes regulate the transcription of Sonic Hedgehog (SHH) and Notch signaling components in neural development
A. Microarrays (Mouse 430 2.0 arrays (Affymetrix) containing 39,000 transcripts) were probed with cRNA generated from RNAs isolated from 3 control and 3 _Brg_-deficient E12.5 telencephalons. Default settings were used to normalize the data and for comparison. Selected genes changed more than two-fold (p<0.01) are shown. Genes that are downregulated in the absence of Brg appear in green and genes that are up-regulated are in red. B. In situ hybridization and immunostaining analyses of transcriptional targets of npBAF complexes in the E13.5 mouse brain. Positive cells are dark brown (coronal sections). Upper panel: Expression of Manic fringe in newly-born neurons requires Brg. VZ: ventricular zone; CP: cortical plate. Middle panel: Repression of TrkB expression in neural progenitors requires Brg. Lower panel: Increased Jagged1 protein levels in _Brg_-deficient basal forebrain. C. BAF45a, BAF53a, Brg and npBAF complexes are present at the promoters of components of the SHH and Notch signaling pathways (P1). P2 regions are ~5 kb upstream. Anti-Brg/Brm, BAF45a and BAF53a antibodies were used for chromatin immunoprecipitation from E13.5 mouse forebrain lysates followed by quantitative PCR. A genomic element of the murine CD4 gene was used as a negative binding control (Chi et al., 2002). The relative binding abundance of each region is presented as percentage of the input DNA material. The data were normalized to the relative abundances of the CD4 P1 region and DNA precipitated by an anti-GFP antibody (negative control) (n=3 independent experiments). Primer sequences are available upon request.
Figure 7.. Repression of BAF45a and BAF53a is essential for neuronal differentiation
A. Left panels: expression of the post-mitotic-specific BAF53b and BAF45b subunits in neural progenitors of the embryonic chicken spinal cord does not interfere with neuronal differentiation. Representative images are shown. Note the high number of (yellow) double positive GFP+ and MNR+ or Islet+ neurons. Right panels: forced expression of the npBAF progenitor-specific BAF45a and BAF53a subunits in post-mitotic neurons is incompatible with neuronal differentiation. Note the low number of (yellow) double-positive GFP+ and MNR+ or Islet+ neurons and the reduced size of the spinal cord on the transfected side. MNR, Motor neuron reactive cells; Isl, Islet-1/2 reactive cells. A quantification of GFP+ /MNR+ or Islet+ double positive neurons is shown. Results are expressed as mean ± SD. **, p<0.01. B. A switch in subunit composition of SWI/SNF-like complexes underlies vertebrate neural development. We propose that Brg and npBAF complexes are required for neural stem cell self-renewal and proliferation (1). The exchange of npBAF and nBAF components is essential for the transition from proliferating neural stem/progenitor cells to post-mitotic neurons (2). BAF complexes are drawn in jigsaw puzzle configuration to denote the apparent fit of the subunits within the complexes. For example, BAF53 and actin directly interact with the ATPase domain of Brg (Zhao et al., 1998). However, the position of the other BAF subunits within the complexes has not been experimentally defined.
Comment in
- A novel model for an older remodeler: the BAF swap in neurogenesis.
Aigner S, Denli AM, Gage FH. Aigner S, et al. Neuron. 2007 Jul 19;55(2):171-3. doi: 10.1016/j.neuron.2007.07.004. Neuron. 2007. PMID: 17640518
Similar articles
- MicroRNA-mediated switching of chromatin-remodelling complexes in neural development.
Yoo AS, Staahl BT, Chen L, Crabtree GR. Yoo AS, et al. Nature. 2009 Jul 30;460(7255):642-6. doi: 10.1038/nature08139. Epub 2009 Jun 28. Nature. 2009. PMID: 19561591 Free PMC article. - The BAF45a/PHF10 subunit of SWI/SNF-like chromatin remodeling complexes is essential for hematopoietic stem cell maintenance.
Krasteva V, Crabtree GR, Lessard JA. Krasteva V, et al. Exp Hematol. 2017 Apr;48:58-71.e15. doi: 10.1016/j.exphem.2016.11.008. Epub 2016 Dec 5. Exp Hematol. 2017. PMID: 27931852 Free PMC article. - MicroRNA-mediated conversion of human fibroblasts to neurons.
Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. Yoo AS, et al. Nature. 2011 Jul 13;476(7359):228-31. doi: 10.1038/nature10323. Nature. 2011. PMID: 21753754 Free PMC article. - Cell fate specification in the mammalian telencephalon.
Guillemot F. Guillemot F. Prog Neurobiol. 2007 Sep;83(1):37-52. doi: 10.1016/j.pneurobio.2007.02.009. Epub 2007 Mar 7. Prog Neurobiol. 2007. PMID: 17517461 Review. - Roles of chromatin remodeling BAF complex in neural differentiation and reprogramming.
Narayanan R, Tuoc TC. Narayanan R, et al. Cell Tissue Res. 2014 Jun;356(3):575-84. doi: 10.1007/s00441-013-1791-7. Epub 2014 Feb 5. Cell Tissue Res. 2014. PMID: 24496512 Review.
Cited by
- Biphasic cell cycle defect causes impaired neurogenesis in down syndrome.
Sharma V, Nehra S, Do LH, Ghosh A, Deshpande AJ, Singhal N. Sharma V, et al. Front Genet. 2022 Oct 12;13:1007519. doi: 10.3389/fgene.2022.1007519. eCollection 2022. Front Genet. 2022. PMID: 36313423 Free PMC article. - Epigenetic control of skeletal muscle regeneration: Integrating genetic determinants and environmental changes.
Giordani L, Puri PL. Giordani L, et al. FEBS J. 2013 Sep;280(17):4014-25. doi: 10.1111/febs.12383. Epub 2013 Jul 15. FEBS J. 2013. PMID: 23745685 Free PMC article. Review. - Targeting specific HATs for neurodegenerative disease treatment: translating basic biology to therapeutic possibilities.
Pirooznia SK, Elefant F. Pirooznia SK, et al. Front Cell Neurosci. 2013 Mar 28;7:30. doi: 10.3389/fncel.2013.00030. eCollection 2013. Front Cell Neurosci. 2013. PMID: 23543406 Free PMC article. - Retinoic acid signaling and neuronal differentiation.
Janesick A, Wu SC, Blumberg B. Janesick A, et al. Cell Mol Life Sci. 2015 Apr;72(8):1559-76. doi: 10.1007/s00018-014-1815-9. Epub 2015 Jan 6. Cell Mol Life Sci. 2015. PMID: 25558812 Free PMC article. Review. - SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh-type medulloblastoma development.
Shi X, Wang Q, Gu J, Xuan Z, Wu JI. Shi X, et al. Oncogene. 2016 Nov 3;35(44):5746-5758. doi: 10.1038/onc.2016.108. Epub 2016 Apr 11. Oncogene. 2016. PMID: 27065321
References
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat.Rev.Neurosci. 2002;3:517–530. - PubMed
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol.Cell. 2000;6:1287–1295. - PubMed
- Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr.Opin.Genet.Dev. 2005;15:185–190. - PubMed
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. - PubMed
- Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS046789/NS/NINDS NIH HHS/United States
- R01 HD055391/HD/NICHD NIH HHS/United States
- R37 NS046789/NS/NINDS NIH HHS/United States
- N01 HV028179/HL/NHLBI NIH HHS/United States
- R01 NS046789/NS/NINDS NIH HHS/United States
- N01-HV-28179/HV/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials