Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells - PubMed (original) (raw)
Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells
Jian Xu et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Recent studies have suggested that, in ES cells, inactive genes encoding early developmental regulators possess bivalent histone modification domains and are therefore poised for activation. However, bivalent domains were not observed at typical tissue-specific genes. Here, we show that windows of unmethylated CpG dinucleotides and putative pioneer factor interactions mark enhancers for at least some tissue-specific genes in ES cells. The unmethylated windows expand in cells that express the gene and contract, disappear, or remain unchanged in nonexpressing tissues. However, in ES cells, they do not always coincide with common histone modifications. Genomic footprinting and chromatin immunoprecipitation demonstrated that transcription factor binding underlies the unmethylated windows at enhancers for the Ptcra and Alb1 genes. After stable integration of premethylated Ptcra enhancer constructs into the ES cell genome, the unmethylated windows readily appeared. In contrast, the premethylated constructs remained fully methylated and silent after introduction into Ptcra-expressing thymocytes. These findings provide initial functional support for a model in which pioneer factor interactions in ES cells promote the assembly of a chromatin structure that is permissive for subsequent activation, and in which differentiated tissues lack the machinery required for gene activation when these ES cell marks are absent. The enhancer marks may therefore represent important features of the pluripotent state.
Conflict of interest statement
Conflict of interest statement: I.L.W. was a member of the scientific advisory board of Amgen, cofounded and is a director of Stem Cells, Inc., and cofounded Cellerant, Inc.
Figures
Fig. 1.
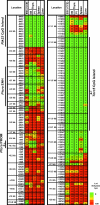
Ptcra methylation in ES cells. Ptcra DNA methylation profiles are shown for the CCE ES cell line, nonstimulated (NS) and PMA/ionomycin-stimulated (PI) thymocytes, spleen, and liver cells. Methylation levels are represented in a gradation of colors: dark green (0–20%), light green (21–40%), yellow (41–60%), orange (61–80%), and red (81–100%). The same data with the ratio values are shown in
SI Fig. 16
.
Fig. 2.
The Ptcra enhancer contains unmethylated windows in numerous tissues. (A) An alignment of the mouse (m) and human (h) Ptcra enhancer core sequences. Previously identified transcription factor binding sites are boxes, and CpG locations are indicated. (B) Methylation profiles for the Ptcra enhancer and promoter in various cells and tissues. Results from a minimum of two experiments are shown for blastocysts, CCE ES cells, thymocytes, BMDM, and sperm cells. Results obtained with the HSC, MPP, CLP, and CMP populations are also described in Attema et al. (40). The same data with the ratio values are shown in
SI Fig. 17
.
Fig. 3.
The Il12b and Alb1 enhancers contain unmethylated windows. (A) Methylation of the Il12 enhancer and promoter in numerous cell types. Results are diagrammed as in Fig. 1. The ratio values are shown in
SI Fig. 8
. (B) CpG methylation in the 300-bp Alb1 enhancer in numerous cell types. (C) Results from five bisulfite sequencing experiments in CCE ES cells. The ratio values are shown in
SI Fig. 9
. (D) DNA sequence of the Alb1 enhancer (−10,566 to −10,865). Transcription factor binding sites are marked above the sequences and CpGs are underlined.
Fig. 4.
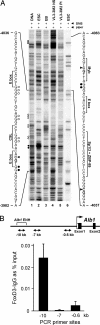
Binding of transcription factors to the Ptcra and Alb1 enhancers in ES cells. (A) In vivo protein–DNA interactions at the Ptcra enhancer region. DMS footprinting was performed with purified CCE ES cell DNA (lane 1), CCE ES cells (lane 2), EB cells (lane 3), Ptcra+ VL3-3M2 thymocytes (lane 4), _Ptcra_− PMA/ionomycin-treated VL3-3M2 cells (lane 5), and CCE ES cells without piperidine cleavage of methylated DNA (lane 6). Nucleotides consistently protected from methylation in ES cells (open circles) and nucleotides exhibiting enhanced methylation (filled circles) are shown. Potential transcription factor binding sites are shown. Filled triangles mark the positions of CpGs (−3,997, −4,042, and −4,080).
SI Fig. 10
shows the results of two additional experiments. (B) Binding of FoxD3 to the Alb1 enhancer in ES cells. ChIP assays were performed with FoxD3 antibodies. Convergent arrows beneath the Alb1 map indicate PCR priming sites. Real-time PCR signals from control IgG ChIPs were subtracted, and the data were expressed as percent input values. Standard deviations from two replicate experiments are shown.
Fig. 5.
ES cells but not thymocytes promote the loss of methylation at a premethylated plasmid. This diagram provides a summary of the results in
SI Fig. 14
. (A) Methylation profile of the premethylated plasmid before transfection. Colors indicate methylation levels as described in the legend to Fig. 1. (B) Methylation profiles for five ES cell clones transfected with the unmethylated Ptcra enhancer-promoter-reporter plasmid and seven ES cell clones transfected with the premethylated plasmid. (C) Methylation profiles for eight VL3-3M2 clones transfected with the unmethylated plasmid and 20 VL3-3M2 clones transfected with the premethylated plasmid. (D) Methylation profiles for four EL4 clones transfected with the unmethylated plasmid and six EL4 clones transfected with the premethylated plasmid.
Similar articles
- Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells.
Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Xu J, et al. Genes Dev. 2009 Dec 15;23(24):2824-38. doi: 10.1101/gad.1861209. Genes Dev. 2009. PMID: 20008934 Free PMC article. - Intragenic CpG islands play important roles in bivalent chromatin assembly of developmental genes.
Lee SM, Lee J, Noh KM, Choi WY, Jeon S, Oh GT, Kim-Ha J, Jin Y, Cho SW, Kim YJ. Lee SM, et al. Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):E1885-E1894. doi: 10.1073/pnas.1613300114. Epub 2017 Feb 21. Proc Natl Acad Sci U S A. 2017. PMID: 28223506 Free PMC article. - Pioneer factors in embryonic stem cells and differentiation.
Smale ST. Smale ST. Curr Opin Genet Dev. 2010 Oct;20(5):519-26. doi: 10.1016/j.gde.2010.06.010. Epub 2010 Jul 16. Curr Opin Genet Dev. 2010. PMID: 20638836 Free PMC article. Review. - The recently identified modifier of murine metastable epialleles, Rearranged L-Myc Fusion, is involved in maintaining epigenetic marks at CpG island shores and enhancers.
Harten SK, Oey H, Bourke LM, Bharti V, Isbel L, Daxinger L, Faou P, Robertson N, Matthews JM, Whitelaw E. Harten SK, et al. BMC Biol. 2015 Mar 26;13:21. doi: 10.1186/s12915-015-0128-2. BMC Biol. 2015. PMID: 25857663 Free PMC article. - Transcriptional Enhancers: Bridging the Genome and Phenome.
Ren B, Yue F. Ren B, et al. Cold Spring Harb Symp Quant Biol. 2015;80:17-26. doi: 10.1101/sqb.2015.80.027219. Epub 2015 Nov 18. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26582789 Review.
Cited by
- Pyrosequencing Evaluation of Widely Available Bisulfite Conversion Methods: Considerations for Application.
Izzi B, Binder AM, Michels KB. Izzi B, et al. Med Epigenet. 2014 Jan 1;2(1):28-36. doi: 10.1159/000358882. Med Epigenet. 2014. PMID: 24944560 Free PMC article. - Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid.
Jin J, Xie X, Xiao Y, Hu H, Zou Q, Cheng X, Sun SC. Jin J, et al. Nat Immunol. 2016 Mar;17(3):259-68. doi: 10.1038/ni.3347. Epub 2016 Jan 25. Nat Immunol. 2016. PMID: 26808229 Free PMC article. - FOXD3 Regulates Pluripotent Stem Cell Potential by Simultaneously Initiating and Repressing Enhancer Activity.
Krishnakumar R, Chen AF, Pantovich MG, Danial M, Parchem RJ, Labosky PA, Blelloch R. Krishnakumar R, et al. Cell Stem Cell. 2016 Jan 7;18(1):104-17. doi: 10.1016/j.stem.2015.10.003. Cell Stem Cell. 2016. PMID: 26748757 Free PMC article. - Tracking the evolution of cancer methylomes.
Krebs AR, Schübeler D. Krebs AR, et al. Nat Genet. 2012 Nov;44(11):1173-4. doi: 10.1038/ng.2451. Nat Genet. 2012. PMID: 23104061
References
- Hochedlinger K, Jaenisch R. Nature. 2006;441:1061–1067. - PubMed
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Genes Dev. 1992;6:705–714. - PubMed
- Jones PA, Takai D. Science. 2001;293:1068–1070. - PubMed
- Reik W, Dean W, Walter J. Science. 2001;293:1089–1093. - PubMed
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Genes Dev. 1996;10:1670–1682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM047903/GM/NIGMS NIH HHS/United States
- P01 DK053074/DK/NIDDK NIH HHS/United States
- GM066367-05/GM/NIGMS NIH HHS/United States
- T32 AI007323/AI/NIAID NIH HHS/United States
- R01 GM47903/GM/NIGMS NIH HHS/United States
- P01 DK53074/DK/NIDDK NIH HHS/United States
- F31 GM066367/GM/NIGMS NIH HHS/United States
- T32 AI07323/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources