Functional dissection of Rab GTPases involved in primary cilium formation - PubMed (original) (raw)
Functional dissection of Rab GTPases involved in primary cilium formation
Shin-Ichiro Yoshimura et al. J Cell Biol. 2007.
Abstract
Primary cilia are sensory structures involved in morphogen signalling during development, liquid flow in the kidney, mechanosensation, sight, and smell (Badano, J.L., N. Mitsuma, P.L. Beales, and N. Katsanis. 2006. Annu. Rev. Genomics Hum. Genet. 7:125-148; Singla, V., and J.F. Reiter. 2006. Science. 313:629-633.). Mutations that affect primary cilia are responsible for several diseases, including neural tube defects, polycystic kidney disease, retinal degeneration, and cancers (Badano et al., 2006; Singla and Reiter, 2006). Primary cilia formation and function requires tight integration of the microtubule cytoskeleton with membrane trafficking (Singla and Reiter, 2006), and this is poorly understood. We show that the Rab GTPase membrane trafficking regulators Rab8a, -17, and -23, and their cognate GTPase-activating proteins (GAPs), XM_037557, TBC1D7, and EVI5like, are involved in primary cilia formation. However, other human Rabs and GAPs are not. Additionally, Rab8a specifically interacts with cenexin/ODF2, a basal body and microtubule binding protein required for cilium biogenesis (Ishikawa, H., A. Kubo, S. Tsukita, and S. Tsukita. 2005. Nat. Cell Biol. 7:517-524), and is the sole Rab enriched at primary cilia. These findings provide a basis for understanding how specific membrane trafficking pathways cooperate with the microtubule cytoskeleton to give rise to the primary cilia.
Figures
Figure 1.
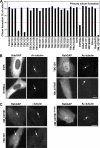
A subset of RabGAPs can block primary cilium formation. (A) hTERT-RPE1 cells expressing human GFP-RabGAPs were induced to form primary cilia by serum starvation and then stained for acetylated tubulin (Ac-tubulin) as a marker for primary cilia. Primary cilium formation was counted (n = 100) and is plotted for a representative series of experiments in the bar graph. The blue line marks the mean extent of cilium formation, and the red line is the 40% cutoff used to assign positive GAPs. TBC1D3 (asterisk) caused reduced cell viability and increased levels of apoptosis; TBC1D12, RUTBC1, RUTBC2, USP6, AK074305, and KIAA0882 gave similar effects and are not shown. (B) Images showing the effects of expressing EVI5like, TBC1D7, and XM_037557 on primary cilia formation. Note the lack of a primary cilium and only residual basal body staining (arrows). EVI5 is shown as a negative control where primary cilium formation is normal (arrow). (C) hTERT-RPE1 cells expressing human GFP-tagged TBC1D7, XM_037557, or the inactive XM_037557R140A mutant were induced to form primary cilia by serum starvation and then stained for γ-tubulin as a marker for the basal body or acetylated tubulin as a marker for the cilium (arrows). DNA was stained with DAPI. Bars, 10 μm.
Figure 2.
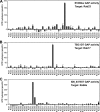
Identification of target Rabs for EVI5like, TBC1D7, and XM_037557. Biochemical GAP assays were performed using a representative set of human Rabs and the candidate RabGAPs EVI5like (A), TBC1D7 (B), and XM_037557 (C). GTP hydrolysis is plotted in pmol/h. The asterisk indicates nonspecific activation of the target Rab. Error bars indicate SD.
Figure 3.
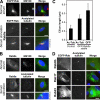
Rab8a is the sole Rab present on primary cilia. (A) hTERT-RPE1 cells expressing human GFP-Rab8a (green) were grown in normal serum or induced to form primary cilia by serum starvation and then stained for acetylated tubulin (red) as a marker for primary cilia. (B) hTERT-RPE1 cells grown in normal serum or induced to form primary cilia by serum starvation were stained for Rab8a (green) and acetylated tubulin (red). (C) The length of primary cilia, defined by acetylated tubulin (Ac-Tub) staining, was measured in control hTERT-RPE1 cells (2.6 ± 0.8 μm) and hTERT-RPE1 cells stably expressing GFP-tagged Rab8a (4.8 ± 0.9 μm). The length of the Rab8a-positive structure (GFP) was also measured (6.7 ± 1.5 μm). These numbers are plotted on the graph with bars to show the standard from the mean (n > 100). (D) hTERT-RPE1 cells expressing Rab8b, -17, and -23 tagged with GFP (green) were induced to form primary cilia by serum starvation and then stained for acetylated tubulin (red) as a marker for primary cilia. DNA was stained with DAPI (blue). Bars, 10 μm.
Figure 4.
Rab8a is required for primary cilium formation. (A and B) hTERT-RPE1 cells treated with control, IFT20, Rab8a, or GMAP210 siRNA duplexes were induced to form primary cilia by serum starvation for 48 h. The cells were fixed and stained for acetylated tubulin (red) as a marker for primary cilia and IFT20 or Rab8a (green). DNA was stained with DAPI (blue). Bar, 10 μm. (C) The number of cells forming primary cilia was counted in cells treated as indicated in the figure. The results are plotted as a bar graph, and the standard error is shown (n = 3; 100 cells per condition).
Figure 5.
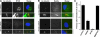
Cenexin 3 is a Rab8a effector at the primary cilium. (A) 13 overlapping fragments of cenexin 3 capable of interacting with Rab8a but not Rab8b were identified using yeast two-hybrid screening of a testis cDNA library. All contain a 20-amino-acid C-terminal region absent from cenexin 1. A schematic of the different cenexin splice variants and these clones is shown in the figure; brown, green, blue, and pink indicate the alternatively spliced regions of cenexin at the N and C termini; interactions are shown by a plus sign and a lack of interaction by a minus sign. Cenexin 3ΔC20, lacking the Rab8 binding domain, did not interact with Rab8a or -8b. These data are summarized in the figure. (B) Binding assays were performed using the Rabs indicated in the figure and the C-terminal domain of cenexin 3 (aa 397–657), or the cenexin 3 C-terminal domain lacking its last 20 amino acids (ΔC20). For cenexin, the entire bound fraction and 20% of the input material are loaded on the gels shown. For Rabs, 1 μg of the input is shown. (C) hTERT-RPE1 cells expressing human FLAG–cenexin 1 or 3 (green) were grown in normal serum or induced to form primary cilia by serum starvation and stained for γ-tubulin or acetylated tubulin (red) as a marker for primary cilia. DNA was stained with DAPI (blue). Bar, 10 μm. (D) The extent of primary cilium formation in hTERT-RPE1 cells expressing cenexin 1, cenexin 3, cenexin 3ΔC20, Cen3R8BD, and Cen3R8DBΔC20 relative to the untreated control was counted and plotted in the bar graph (n = 3; 100 cells per condition). Error bars indicate SD. (E) A model for primary cilium formation. Microtubules and the basal body are shown in green, and arrows mark the Rab-regulated membrane trafficking pathways. The cilia membrane domain defined by Rab8a is marked in blue, and circular red arrows indicate the points of GAP regulation.
Similar articles
- Analysis of Rab GTPase and GTPase-activating protein function at primary cilia.
Yoshimura S, Haas AK, Barr FA. Yoshimura S, et al. Methods Enzymol. 2008;439:353-64. doi: 10.1016/S0076-6879(07)00426-0. Methods Enzymol. 2008. PMID: 18374177 - Molecular dissection of ODF2/Cenexin revealed a short stretch of amino acids necessary for targeting to the centrosome and the primary cilium.
Hüber D, Geisler S, Monecke S, Hoyer-Fender S. Hüber D, et al. Eur J Cell Biol. 2008 Mar;87(3):137-46. doi: 10.1016/j.ejcb.2007.10.004. Epub 2008 Jan 2. Eur J Cell Biol. 2008. PMID: 18171590 - A comprehensive analysis of Rab GTPases reveals a role for Rab34 in serum starvation-induced primary ciliogenesis.
Oguchi ME, Okuyama K, Homma Y, Fukuda M. Oguchi ME, et al. J Biol Chem. 2020 Sep 4;295(36):12674-12685. doi: 10.1074/jbc.RA119.012233. Epub 2020 Jul 15. J Biol Chem. 2020. PMID: 32669361 Free PMC article. - Rabs and other small GTPases in ciliary transport.
Lim YS, Chua CE, Tang BL. Lim YS, et al. Biol Cell. 2011 May;103(5):209-21. doi: 10.1042/BC20100150. Biol Cell. 2011. PMID: 21488838 Review. - Rab GTPases in cilium formation and function.
Blacque OE, Scheidel N, Kuhns S. Blacque OE, et al. Small GTPases. 2018 Mar 4;9(1-2):76-94. doi: 10.1080/21541248.2017.1353847. Epub 2017 Oct 26. Small GTPases. 2018. PMID: 29072526 Free PMC article. Review.
Cited by
- The Msb3/Gyp3 GAP controls the activity of the Rab GTPases Vps21 and Ypt7 at endosomes and vacuoles.
Lachmann J, Barr FA, Ungermann C. Lachmann J, et al. Mol Biol Cell. 2012 Jul;23(13):2516-26. doi: 10.1091/mbc.E11-12-1030. Epub 2012 May 16. Mol Biol Cell. 2012. PMID: 22593206 Free PMC article. - Molecular assemblies that control rhodopsin transport to the cilia.
Deretic D, Wang J. Deretic D, et al. Vision Res. 2012 Dec 15;75:5-10. doi: 10.1016/j.visres.2012.07.015. Epub 2012 Aug 7. Vision Res. 2012. PMID: 22892112 Free PMC article. Review. - Transcription Activation of Rab8A by PEA3 Augments Progression of Esophagus Cancer by Activating the Wnt/_β_-Catenin Signaling Pathway.
Cai S, Liu X, Gao W, Ye L, Zhong Q, Zhang X. Cai S, et al. Dis Markers. 2023 Feb 13;2023:8143581. doi: 10.1155/2023/8143581. eCollection 2023. Dis Markers. 2023. PMID: 36815135 Free PMC article. - Rab10 associates with primary cilia and the exocyst complex in renal epithelial cells.
Babbey CM, Bacallao RL, Dunn KW. Babbey CM, et al. Am J Physiol Renal Physiol. 2010 Sep;299(3):F495-506. doi: 10.1152/ajprenal.00198.2010. Epub 2010 Jun 24. Am J Physiol Renal Physiol. 2010. PMID: 20576682 Free PMC article. - Rab35 controls cilium length, function and membrane composition.
Kuhns S, Seixas C, Pestana S, Tavares B, Nogueira R, Jacinto R, Ramalho JS, Simpson JC, Andersen JS, Echard A, Lopes SS, Barral DC, Blacque OE. Kuhns S, et al. EMBO Rep. 2019 Oct 4;20(10):e47625. doi: 10.15252/embr.201847625. Epub 2019 Aug 21. EMBO Rep. 2019. PMID: 31432619 Free PMC article.
References
- Badano, J.L., N. Mitsuma, P.L. Beales, and N. Katsanis. 2006. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7:125–148. - PubMed
- Behnia, R., and S. Munro. 2005. Organelle identity and the signposts for membrane traffic. Nature. 438:597–604. - PubMed
- Caspary, T., C.E. Larkins, and K.V. Anderson. 2007. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 12:767–778. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases