The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia - PubMed (original) (raw)
The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia
Benjamin J Thompson et al. J Exp Med. 2007.
Abstract
Recent studies have shown that activating mutations of NOTCH1 are responsible for the majority of T cell acute lymphoblastic leukemia (T-ALL) cases. Most of these mutations truncate its C-terminal domain, a region that is important for the NOTCH1 proteasome-mediated degradation. We report that the E3 ligase FBW7 targets NOTCH1 for ubiquitination and degradation. Our studies map in detail the amino acid degron sequence required for NOTCH1-FBW7 interaction. Furthermore, we identify inactivating FBW7 mutations in a large fraction of human T-ALL lines and primary leukemias. These mutations abrogate the binding of FBW7 not only to NOTCH1 but also to the two other characterized targets, c-Myc and cyclin E. The majority of the FBW7 mutations were present during relapse, and they were associated with NOTCH1 HD mutations. Interestingly, most of the T-ALL lines harboring FBW7 mutations were resistant to gamma-secretase inhibitor treatment and this resistance appeared to be related to the stabilization of the c-Myc protein. Our data suggest that FBW7 is a novel tumor suppressor in T cell leukemia, and implicate the loss of FBW7 function as a potential mechanism of drug resistance in T-ALL.
Figures
Figure 1.
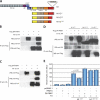
Physical and functional association between FBW7 and NOTCH1. (A) Schematic depiction of the NOTCH1 (N1) protein domains, and N1-IC WT (N1-ICWT) and 82 and 162 ΔPEST mutants (N1-ICΔ82 and N1-ICΔ162). Asterisk denotes L1601P mutation. The FBW7 degron is highlighted in gray. LNR, LIN-Notch repeat domain; RAM, RBJ-κ–associated module; TAD, transcriptional activation domain; PEST, P-E-S-T–rich domain; HD, heterodimerization domain. (B and C) Coimmunoprecipitation experiments using anti-Flag beads or anti-HA beads, showing interaction between DN FBW7 and N1-ICWT, but DN FBW7 (D) does not interact with N1-ICΔ82 and N1-ICΔ162. (E) Cotransfection with FBW7α repressed N1L1601P (P < 0.02), but not N1L1601P-Δ82, in reporter assays using a CSL luciferase reporter. pCDNA3.1 represents the control, “empty” expression vector. Error bars represent the SD of duplicate wells.
Figure 2.
Mutational analysis of the FBW7 NOTCH1 degron. (A) Coimmunoprecipitation experiment using threonine mutants of N1-IC (N1-ICT2133A, N1-ICT2467A, N1-ICT2484A, and N1-ICT2512A) and DN FBW7. Only N1-ICT2512A was unable to coimmunoprecipitate with DN FBW7. (B) Cell lysates from Bosc23 cells transfected with N1L1601P or N1L1601P-T2512A and FBW7α were used for luciferase reporter assays. N1L1601P-T2512A was not repressed by FBW7α. (C) Cell lysates from HeLa cells transfected with full-length N1 mutants were used for luciferase reporter assays. N1L1601P-Δ82 and N1L1601P-T2512A induced a 15- and 5-fold increase in transcriptional activity, respectively, compared with N1L1601P. (D) In vivo ubiquitination assay using Bosc23 cells transfected with N1-ICWT, N1-ICΔ82, or N1-ICT2512A with or without SCFFBW7α (SKP1, CUL1, ROC1, and FBW7α) and Ub-HA. SCFFBW7α enhanced polyubiquitination of N1-ICWT, but not of N1-ICΔ82 or N1-ICT2512A. (E) A negative charge is required at position 2516 (T+4). Interaction with DN FBW7 is partially preserved with N1-ICE2516D, but abolished with N1-ICE2516A. (F) Mutation of two CDK8 phosphorylation sites prevents N1-IC interaction with FBW7. A mutant lacking both S2514 and S2517 (N1-ICS2514A-S2517A) did not coimmunoprecipitate with DN FBW7. Error bars represent the SD of duplicate wells.
Figure 3.
FBW7 mutations in T-ALL preclude interaction with NOTCH1, c-Myc, and cyclin E. (A) Schematic depiction of the positions of the FBW7 binding pocket and DN FBW7. (B) Chromatograms showing mutations of FBW7 R465 and R505 found in CEM and P12-ICHIKAWA T-ALL cell lines. DND41 cells express WT FBW7. Arrows indicate mutated nucleotides. (C) Coimmunoprecipitation experiments demonstrating the inability of DN FBW7R465C, DN FBW7R479Q, and DN FBW7R505C to interact with N1-IC. (D and E) The same DN FBW7 mutants as in C do not interact with c-Myc and cyclin E. Asterisk in E indicates a nonspecific band.
Figure 4.
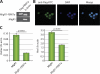
Restoration of FBW7 expression in CEM cells (FBW7MUT) suppresses NOTCH1 target genes. (A and B) Efficient restoration of FBW7WT in CEM T-ALL cells after transduction with a Flag-FBW7α–expressing retrovirus (MigR1-FBW7α). Cells were infected with empty MigR1 vector as a control. (A) RT-PCR amplified Flag-FBW7α only in cells infected with MigR1-FBW7α. (B) Immunofluorescence staining of FACS-sorted CEM cells infected with MigR1-FBW7α using FITC-conjugated anti-Flag antibody shows nuclear localization of the construct. (C) Real-time RT-PCR was used to quantify the expression of the NOTCH1 target genes, DELTEX1, and HES1 in CEM cells infected as in A Infection with MigR1-FBW7α suppressed the two genes (P < 0.0001 for both). Error bars represent the SD of triplicate wells.
Figure 5.
FBW7 mutations as a mechanism of resistance to γ-secretase inhibition. (A) T-ALL cell lines were treated with MG132 (20 μM for 6 h) and analyzed for expression of c-Myc and actin. Treatment with MG132 resulted in accumulation of c-Myc in KOPTK cells, which express WT FBW7, but not in Be13 and P12-ICHIKAWA cells, which express no and mutant FBW7, respectively. (B) Expression of N1-IC and c-Myc after treatment with Compound E (CompE) for 1–4 d. c-Myc expression declines only in KOPTK1 cells, which are sensitive to this treatment, but not in Be13 and P12-ICHIKAWA cells, which are resistant. N1-IC expression decreased in all of the cell lines, reflecting inhibition of γ-secretase–mediated cleavage of NOTCH1. Actin expression was used as a loading control for both A and B.
References
- Grabher, C., H. von Boehmer, and A.T. Look. 2006. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 6:1–13. - PubMed
- Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. - PubMed
- Maillard, I., T. Fang, and W.S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the notch pathway. Annu. Rev. Immunol. 23:945–974. - PubMed
- Rothenberg, E.V., and T. Taghon. 2005. Molecular genetics of T cell development. Annu. Rev. Immunol. 23:601–649. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases