Heterologous amyloid seeding: revisiting the role of acetylcholinesterase in Alzheimer's disease - PubMed (original) (raw)
Heterologous amyloid seeding: revisiting the role of acetylcholinesterase in Alzheimer's disease
Létitia Jean et al. PLoS One. 2007.
Abstract
Neurodegenerative diseases associated with abnormal protein folding and ordered aggregation require an initial trigger which may be infectious, inherited, post-inflammatory or idiopathic. Proteolytic cleavage to generate vulnerable precursors, such as amyloid-beta peptide (Abeta) production via beta and gamma secretases in Alzheimer's Disease (AD), is one such trigger, but the proteolytic removal of these fragments is also aetiologically important. The levels of Abeta in the central nervous system are regulated by several catabolic proteases, including insulysin (IDE) and neprilysin (NEP). The known association of human acetylcholinesterase (hAChE) with pathological aggregates in AD together with its ability to increase Abeta fibrilization prompted us to search for proteolytic triggers that could enhance this process. The hAChE C-terminal domain (T40, AChE(575-614)) is an exposed amphiphilic alpha-helix involved in enzyme oligomerisation, but it also contains a conformational switch region (CSR) with high propensity for conversion to non-native (hidden) beta-strand, a property associated with amyloidogenicity. A synthetic peptide (AChE(586-599)) encompassing the CSR region shares homology with Abeta and forms beta-sheet amyloid fibrils. We investigated the influence of IDE and NEP proteolysis on the formation and degradation of relevant hAChE beta-sheet species. By combining reverse-phase HPLC and mass spectrometry, we established that the enzyme digestion profiles on T40 versus AChE(586-599), or versus Abeta, differed. Moreover, IDE digestion of T40 triggered the conformational switch from alpha- to beta-structures, resulting in surfactant CSR species that self-assembled into amyloid fibril precursors (oligomers). Crucially, these CSR species significantly increased Abeta fibril formation both by seeding the energetically unfavorable formation of amyloid nuclei and by enhancing the rate of amyloid elongation. Hence, these results may offer an explanation for observations that implicate hAChE in the extent of Abeta deposition in the brain. Furthermore, this process of heterologous amyloid seeding by a proteolytic fragment from another protein may represent a previously underestimated pathological trigger, implying that the abundance of the major amyloidogenic species (Abeta in AD, for example) may not be the only important factor in neurodegeneration.
Conflict of interest statement
Competing Interests: Synaptica Ltd holds patents on the use of the AChE586-599 peptide and related peptides within T40 as potential biomarkers for neurodegenerative disease. The University of Oxford holds patents on the method of plate-based surface tension measurement.
Figures
Figure 1. Secondary structure propensity of T40 as predicted by hidden β-propensity method (available at http://opal.umdnj.edu).
Propensities for helices (red squares), β-strands (blue squares) and random coil (green squares) are presented numerically using a 0-1 scale, with low values indicating zero to low propensity and high values indicating high propensity to near certainty.
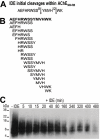
Figure 2. T40-degrading activity of IDE.
(A) IDE degrades non-amyloidogenic and monomeric T40. 16 µM T40 was incubated with or without 22 nM IDE (37°C) and the reaction stopped as indicated. (B) Specificity of IDE activity. 16 µM T40 was incubated (37°C, 90 min) with no IDE, with 22 nM IDE or with 2–16 µM insulin, 2–16 µM IgG, 1 and 10 mM 1,10-phenanthroline, or 0.3 and 3 µL methanol. For (A) and (B), digestion products were resolved (10% Tris-Tricine SDS-PAGE), electro-blotted onto nitrocellulose and probed with KD69 (specific for the T40). Marker proteins are indicated. Arrows indicate the positions of T40 monomers and dimers. (C) 60 µM T40 was incubated with or without 50 nM IDE (37°C) and products separated by RP-HPLC (peaks annotated a–h). (D) Positions of IDE initial cleavages (arrows) within T40 (2 min digestion). (E) Identity of the major peptides (italics) and of CSR species in peaks a–h, analyzed by MS. (F) The relative abundance of CSR species was determined with reference to an internal standard on the MS spectra and is displayed as arbitrary units. ‘+’, 1 to 25 arbitrary units; ‘++’, 26 to 100; ‘+++’, 101 to 250; and ‘++++’, >250. Potential precursors of CSR species are italicized and shown in bold. ‘*’ indicates CSR species that were not present after a 2 hour digestion of T40 by IDE.
Figure 3. IDE degrades both monomeric and oligomeric forms of AChE586-599.
(A) Positions of IDE initial cleavages (arrows) within AChE586-599. (B) Cleavage map after complete IDE digestion of AChE586-599. 63 µM AChE586-599 was incubated with or without 45 nM IDE (A) or 273 nM IDE (B)(37°C, 30 min) and RP-HPLC peaks analyzed by MS. (C) IDE degrades AChE586-599 oligomers. AChE586-599 oligomers (16 µM) cross-linked by photo-induced cross-linking were incubated with or without 16.3 nM IDE (37°C). Digestion products were resolved (16.5% Tris-Tricine SDS-PAGE), electro-blotted onto nitrocellulose and probed with Mab 105A (specific for AChE586-599 in β-sheet conformation). Marker proteins are indicated.
Figure 4. NEP preferentially degrades monomeric forms of AChE586-599.
(A) 60 µM of either substance P or T40 were incubated with or without 1.2 µM NEP (37°C, 4 hours) and the digestion mixture subjected to RP-HPLC. (B) Positions of NEP initial cleavages (arrows) within AChE586-599. (C) Cleavage map after complete NEP digestion of AChE586-599. 40 µM AChE586-599 was incubated with 772 nM NEP (30 min (B) or 4 hours (C), 37°C) and RP-HPLC peaks analyzed by MS. (D) NEP degrades AChE586-599 oligomers. AChE586-599 oligomers (14.8 µM) cross-linked by photo-induced cross-linking were incubated with or without 285 nM NEP (37°C). Digestion products were analyzed as described in Figure 3C. A_rrows_ indicate two digested AChE586-599 oligomers. (E) T40/IDE products are substrates for NEP. 60 µM T40 was incubated with 50 nM IDE (37°C, 30 min) and products separated by RP-HPLC. Peaks e–f (see Figure. 2C) were lyophilized, incubated with 772 nM NEP (37°C, 2 hours) and subjected to RP-HPLC.
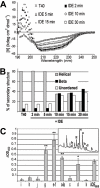
Figure 5. Conformation and surfactant properties of CSR species generated from T40 by IDE.
(A) and (B) T40/IDE digestion triggers a switch to β-structure. T40 CD spectra (250 to 190 nm) before and after addition of IDE (A) and percentage of secondary structures (B). (C) CSR species are surface-active. T40/IDE digest (30 min) was subjected to RP-HPLC. Peaks (a–k, as annotated in the inset) were lyophilized, re-suspended in 200 mM sodium acetate pH 3 and surface tension measured before and after neutralization (1M NaH2PO4, pH7.2). Surface tension calculations were as described in Methods. ‘*’ p<0.05, and ‘**’ p<0.007.
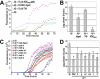
Figure 6. T40/IDE digestion products promote Aβ fibrilization.
15 µM Aβ was incubated with 165 µM ThT, with or without 15 µM T40, 15 µM AChE586-599 seeds, RP-HPLC isolated T40/IDE total digest or individual peaks (a–h, see Figure. 1C)(∼10 µM starting T40). Changes in ThT fluorescence were monitored (A and C) with the lag phase of Aβ fibrilization depicted (B and D). ‘*’ p<0.006 (B) and p<0.05 (D). Control experiments showed that there was no carry over of IDE activity in the T40/IDE digest under the sample preparation conditions (RP-HPLC and lyophilizations).
Figure 7. T40/IDE digestion products form amyloid protofibrils.
Electron micrographs of negatively stained T40/IDE total digest showing spherical (A) annular (B, I–II) and beaded (C, III) protofibrils. The bar represents 10 nm.
Similar articles
- In vivo localization of human acetylcholinesterase-derived species in a β-sheet conformation at the core of senile plaques in Alzheimer's disease.
Jean L, Brimijoin S, Vaux DJ. Jean L, et al. J Biol Chem. 2019 Apr 19;294(16):6253-6272. doi: 10.1074/jbc.RA118.006230. Epub 2019 Feb 20. J Biol Chem. 2019. PMID: 30787102 Free PMC article. - Green Tea Seed Isolated Theasaponin E1 Ameliorates AD Promoting Neurotoxic Pathogenesis by Attenuating Aβ Peptide Levels in SweAPP N2a Cells.
Khan MI, Shin JH, Kim MY, Shin TS, Kim JD. Khan MI, et al. Molecules. 2020 May 16;25(10):2334. doi: 10.3390/molecules25102334. Molecules. 2020. PMID: 32429462 Free PMC article. - Aggregation and catabolism of disease-associated intra-Abeta mutations: reduced proteolysis of AbetaA21G by neprilysin.
Betts V, Leissring MA, Dolios G, Wang R, Selkoe DJ, Walsh DM. Betts V, et al. Neurobiol Dis. 2008 Sep;31(3):442-50. doi: 10.1016/j.nbd.2008.06.001. Epub 2008 Jun 17. Neurobiol Dis. 2008. PMID: 18602473 Free PMC article. - Alzheimer's disease.
De-Paula VJ, Radanovic M, Diniz BS, Forlenza OV. De-Paula VJ, et al. Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review. - Impact of Insulin Degrading Enzyme and Neprilysin in Alzheimer's Disease Biology: Characterization of Putative Cognates for Therapeutic Applications.
Jha NK, Jha SK, Kumar D, Kejriwal N, Sharma R, Ambasta RK, Kumar P. Jha NK, et al. J Alzheimers Dis. 2015;48(4):891-917. doi: 10.3233/JAD-150379. J Alzheimers Dis. 2015. PMID: 26444774 Review.
Cited by
- Conformational preferences of a 14-residue fibrillogenic peptide from acetylcholinesterase.
Vijayan R, Biggin PC. Vijayan R, et al. Biochemistry. 2010 May 4;49(17):3678-84. doi: 10.1021/bi1001807. Biochemistry. 2010. PMID: 20356043 Free PMC article. - In vivo localization of human acetylcholinesterase-derived species in a β-sheet conformation at the core of senile plaques in Alzheimer's disease.
Jean L, Brimijoin S, Vaux DJ. Jean L, et al. J Biol Chem. 2019 Apr 19;294(16):6253-6272. doi: 10.1074/jbc.RA118.006230. Epub 2019 Feb 20. J Biol Chem. 2019. PMID: 30787102 Free PMC article. - Can Activation of Acetylcholinesterase by β-Amyloid Peptide Decrease the Effectiveness of Cholinesterase Inhibitors?
Zueva IV, Vasilieva EA, Gaynanova GA, Moiseenko AV, Burtseva AD, Boyko KM, Zakharova LY, Petrov KA. Zueva IV, et al. Int J Mol Sci. 2023 Nov 16;24(22):16395. doi: 10.3390/ijms242216395. Int J Mol Sci. 2023. PMID: 38003588 Free PMC article. - Advances in the pathogenesis of Alzheimer's disease: a re-evaluation of amyloid cascade hypothesis.
Dong S, Duan Y, Hu Y, Zhao Z. Dong S, et al. Transl Neurodegener. 2012 Sep 21;1(1):18. doi: 10.1186/2047-9158-1-18. Transl Neurodegener. 2012. PMID: 23210692 Free PMC article. - The human acetylcholinesterase C-terminal T30 peptide activates neuronal growth through alpha 7 nicotinic acetylcholine receptors and the mTOR pathway.
Graur A, Sinclair P, Schneeweis AK, Pak DT, Kabbani N. Graur A, et al. Sci Rep. 2023 Jul 15;13(1):11434. doi: 10.1038/s41598-023-38637-1. Sci Rep. 2023. PMID: 37454238 Free PMC article.
References
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid β-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. - PubMed
- Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat Cell Biol. 2004;6:1054–1061. - PubMed
- Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, et al. Metabolic regulation of brain Aβ by neprilysin. Science. 2001;292:1550–1552. - PubMed
- Kurochkin IV. Insulin-degrading enzyme: embarking on amyloid destruction. Trends Biochem Sci. 2001;26:421–425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical