Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis - PubMed (original) (raw)
Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis
Yurii S Aulchenko et al. Genetics. 2007 Sep.
Abstract
For pedigree-based quantitative trait loci (QTL) association analysis, a range of methods utilizing within-family variation such as transmission-disequilibrium test (TDT)-based methods have been developed. In scenarios where stratification is not a concern, methods exploiting between-family variation in addition to within-family variation, such as the measured genotype (MG) approach, have greater power. Application of MG methods can be computationally demanding (especially for large pedigrees), making genomewide scans practically infeasible. Here we suggest a novel approach for genomewide pedigree-based quantitative trait loci (QTL) association analysis: genomewide rapid association using mixed model and regression (GRAMMAR). The method first obtains residuals adjusted for family effects and subsequently analyzes the association between these residuals and genetic polymorphisms using rapid least-squares methods. At the final step, the selected polymorphisms may be followed up with the full measured genotype (MG) analysis. In a simulation study, we compared type 1 error, power, and operational characteristics of the proposed method with those of MG and TDT-based approaches. For moderately heritable (30%) traits in human pedigrees the power of the GRAMMAR and the MG approaches is similar and is much higher than that of TDT-based approaches. When using tabulated thresholds, the proposed method is less powerful than MG for very high heritabilities and pedigrees including large sibships like those observed in livestock pedigrees. However, there is little or no difference in empirical power of MG and the proposed method. In any scenario, GRAMMAR is much faster than MG and enables rapid analysis of hundreds of thousands of markers.
Figures
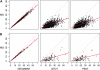
Figure 1.—
Scatterplot of the χ2-statistics coming from the measured-genotype (MG) approach vs. χ2-statistics coming from GRAMMAR, QTDT, and FBAT. The model assumes heritability of 0.28 and a QTL explaining 0.02 of the total variance. Results of analysis of data from 337 sib-trios (A) and ERF pedigree (B) are presented. The dotted line represents a slope of unity.
Similar articles
- Transmission disequilibrium test for quantitative trait loci detection in livestock populations.
Kolbehdari D, Jansen GB, Schaeffer LR, Allen BO. Kolbehdari D, et al. J Anim Breed Genet. 2006 Jun;123(3):191-7. doi: 10.1111/j.1439-0388.2006.00579.x. J Anim Breed Genet. 2006. PMID: 16706924 - Family-based association tests for genomewide association scans.
Chen WM, Abecasis GR. Chen WM, et al. Am J Hum Genet. 2007 Nov;81(5):913-26. doi: 10.1086/521580. Epub 2007 Sep 18. Am J Hum Genet. 2007. PMID: 17924335 Free PMC article. - Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites.
Abney M, Ober C, McPeek MS. Abney M, et al. Am J Hum Genet. 2002 Apr;70(4):920-34. doi: 10.1086/339705. Epub 2002 Mar 4. Am J Hum Genet. 2002. PMID: 11880950 Free PMC article. - Why breeding values estimated using familial data should not be used for genome-wide association studies.
Ekine CC, Rowe SJ, Bishop SC, de Koning DJ. Ekine CC, et al. G3 (Bethesda). 2014 Feb 19;4(2):341-7. doi: 10.1534/g3.113.008706. G3 (Bethesda). 2014. PMID: 24362310 Free PMC article. - Quantitative Trait Loci Identification by Estimating the Genetic Model based on the Extremal Samples.
Yang Z, Yang Y, Xu XS, Yuan M. Yang Z, et al. Curr Genomics. 2021 Dec 30;22(5):363-372. doi: 10.2174/1389202922666210625161602. Curr Genomics. 2021. PMID: 35283669 Free PMC article. Review.
Cited by
- High-throughput analysis of epistasis in genome-wide association studies with BiForce.
Gyenesei A, Moody J, Semple CA, Haley CS, Wei WH. Gyenesei A, et al. Bioinformatics. 2012 Aug 1;28(15):1957-64. doi: 10.1093/bioinformatics/bts304. Epub 2012 May 21. Bioinformatics. 2012. PMID: 22618535 Free PMC article. - Effect of the prior distribution of SNP effects on the estimation of total breeding value.
Nadaf J, Riggio V, Yu TP, Pong-Wong R. Nadaf J, et al. BMC Proc. 2012 May 21;6 Suppl 2(Suppl 2):S6. doi: 10.1186/1753-6561-6-S2-S6. Epub 2012 May 21. BMC Proc. 2012. PMID: 22640730 Free PMC article. - Dissecting Complex Traits Using Omics Data: A Review on the Linear Mixed Models and Their Application in GWAS.
Alamin M, Sultana MH, Lou X, Jin W, Xu H. Alamin M, et al. Plants (Basel). 2022 Nov 28;11(23):3277. doi: 10.3390/plants11233277. Plants (Basel). 2022. PMID: 36501317 Free PMC article. Review. - Genome-wide association analyses of quantitative traits: the GAW16 experience.
Ghosh S. Ghosh S. Genet Epidemiol. 2009;33 Suppl 1(Suppl 1):S13-8. doi: 10.1002/gepi.20466. Genet Epidemiol. 2009. PMID: 19924711 Free PMC article. - Identification of QTLs for wheat heading time across multiple-environments.
Benaouda S, Dadshani S, Koua P, Léon J, Ballvora A. Benaouda S, et al. Theor Appl Genet. 2022 Aug;135(8):2833-2848. doi: 10.1007/s00122-022-04152-6. Epub 2022 Jul 1. Theor Appl Genet. 2022. PMID: 35776141 Free PMC article.
References
- Aulchenko, Y. S., S. Ripke, A. Isaacs and C. M. van Duijn, 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics (in press). - PubMed
- Blangero, J., 2004. Localization and identification of human quantitative trait loci: king harvest has surely come. Curr. Opin. Genet. Dev. 14: 233–240. - PubMed
- Boerwinkle, E., R. Chakraborty and C. F. Sing, 1986. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann. Hum. Genet. 50(Pt. 2): 181–194. - PubMed
- Bourgain, C., and E. Genin, 2005. Complex trait mapping in isolated populations: Are specific statistical methods required? Eur. J. Hum. Genet. 13: 698–706. - PubMed
Publication types
MeSH terms
Grants and funding
- BBS/E/R/00001604/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/R/00001606/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources