Regulation of photoreceptor gene expression by Crx-associated transcription factor network - PubMed (original) (raw)
Review
Regulation of photoreceptor gene expression by Crx-associated transcription factor network
Anne K Hennig et al. Brain Res. 2008.
Abstract
Rod and cone photoreceptors in the mammalian retina are special types of neurons that are responsible for phototransduction, the first step of vision. Development and maintenance of photoreceptors require precisely regulated gene expression. This regulation is mediated by a network of photoreceptor transcription factors centered on Crx, an Otx-like homeodomain transcription factor. The cell type (subtype) specificity of this network is governed by factors that are preferentially expressed by rods or cones or both, including the rod-determining factors neural retina leucine zipper protein (Nrl) and the orphan nuclear receptor Nr2e3; and cone-determining factors, mostly nuclear receptor family members. The best-documented of these include thyroid hormone receptor beta2 (Tr beta2), retinoid related orphan receptor Ror beta, and retinoid X receptor Rxr gamma. The appropriate function of this network also depends on general transcription factors and cofactors that are ubiquitously expressed, such as the Sp zinc finger transcription factors and STAGA co-activator complexes. These cell type-specific and general transcription regulators form complex interactomes; mutations that interfere with any of the interactions can cause photoreceptor development defects or degeneration. In this manuscript, we review recent progress on the roles of various photoreceptor transcription factors and interactions in photoreceptor subtype development. We also provide evidence of auto-, para-, and feedback regulation among these factors at the transcriptional level. These protein-protein and protein-promoter interactions provide precision and specificity in controlling photoreceptor subtype-specific gene expression, development, and survival. Understanding these interactions may provide insights to more effective therapeutic interventions for photoreceptor diseases.
Figures
Figure 1. Schematic diagram of photoreceptor-specific transcription factors
The domain structures of photoreceptor-specific transcription factors discussed in this paper are presented in scale. Conserved domains for classifying families of transcription factors are indicated in black, other regions of homology conserved among different family members are indicated by stippling. Functional regions are indicated above the box representing the factor; sites of mutations discussed in the text are indicated by arrowheads below the box. N- and C-terminals are indicated, and the number below the C-terminal end indicates the number of amino acids in the human protein. HOMEO, homeodomain; b, basic domain; L Zipper, leucine zipper domain; Zn F, zinc finger domain.
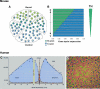
Figure 2. Distribution of cones and opsin expression in mouse and human retina
A. In the mouse, cones are scattered throughout the retina, with M-cones (green) predominating in the dorsal retina and S-cones (blue) predominating in the ventral retina. Many cones express both opsins. B. M-opsin and S-opsin are expressed in complementary dorsal (D) to ventral (V) gradients across the mouse retina, likely in response to a gradient of thyroid hormone (TH), which is established between P4 and P10, and is highest in the dorsal retina as indicated. C. This graph shows the spatial density of rods (blue) and cones (orange) in a horizontal strip of human retina across the fovea (centered at position 0) and optic disc. Cones are concentrated in the fovea, and rods are excluded from this region. Elsewhere in the retina, rods predominate and cones are sparse. D. Within the fovea, cones expressing either green or red opsin predominate, with cones expression blue opsin found sparsely around the peripheral fovea region. Human cones only express a single type of opsin. Panels A and B are from (Applebury et al., 2000), reprinted by permission from Cell Press, with TH gradient added from (Roberts et al., 2006). Panel C is from (Rodieck, 1998), pg. 43, reprinted by permission from Sinauer Associates, Inc. Panel D is from (Cepko, 2000), Figure 2, reprinted by permission from Macmillan Publishers, Ltd: Nature 24: 99−100, copyright 2000.
Figure 3. ChIP analysis demonstrating that network transcription factors bind to their own and each other's promoters
Antibodies against Crx, Nrl, Nr2e3, Trβ2, and NeuroD1 were used to immunoprecipitate the bound chromatin fragments from wild-type (WT), Crx−/−, Nrl−/−, or Nr2e3rd7/rd7 (“Nr2e3−/−“) retinae. Primers specific to the promoter regions of the genes listed on the left [(Peng and Chen, 2005); Table 3] were used to detect the presence of the candidate promoter regions in the immunoprecipitates by PCR. A band indicates that the transcription factor recognized by the immunoprecipitating antibody is bound to the promoter region of the indicated regulator or target gene. Target genes examined include S-opsin (Sop), M-opsin (Mop), rhodopsin (Rho), and interphotoreceptor binding protein (Rbp3), all of which are expressed in photoreceptors. GluR6, which is expressed in bipolar cells but not photoreceptors, serves as a control for photoreceptor specificity. In addition, PCR reactions using primers against DNA sequences immediately 3’ of each gene gave no bands (data not shown), confirming regulatory region-specific binding. Control immunoprecipitates using purified non-specific rabbit or goat IgG yielded no specific promoter sequences from WT (second lanes from the right) or knockout (data not shown) mice. Samples of retina homogenates (“input”) from WT (far right lanes) and knockout (data not shown) mice serve as positive controls for PCR.
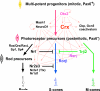
Figure 4. Model for transcription factor network regulation of photoreceptor subtype development
Photoreceptor subtypes develop from photoreceptor precursors derived from multi-potent progenitors via three major pathways (thick arrows). Photoreceptor transcription factors that play a major role in this process are listed based on their epistatic relationship as determined by in vivo and/or in vitro functional studies. Thin lines show protein-promoter interactions; solid lines show interactions reported here and/or previously; dotted lines are from unpublished data. Arrows indicate positive regulation, while blocked lines represent inhibition/suppression. Absence of lines indicates that the relationship remains to be determined.
Similar articles
- In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development.
Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. Cheng H, et al. Hum Mol Genet. 2006 Sep 1;15(17):2588-602. doi: 10.1093/hmg/ddl185. Epub 2006 Jul 25. Hum Mol Genet. 2006. PMID: 16868010 Free PMC article. - The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes.
Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. Peng GH, et al. Hum Mol Genet. 2005 Mar 15;14(6):747-64. doi: 10.1093/hmg/ddi070. Epub 2005 Feb 2. Hum Mol Genet. 2005. PMID: 15689355 - Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors.
Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Cheng H, et al. Hum Mol Genet. 2004 Aug 1;13(15):1563-75. doi: 10.1093/hmg/ddh173. Epub 2004 Jun 9. Hum Mol Genet. 2004. PMID: 15190009 - [Transcriptional regulation of retinal photoreceptor cell development].
Inoue T, Nishida A, Furukawa T. Inoue T, et al. Tanpakushitsu Kakusan Koso. 2004 Aug;49(10):1413-20. Tanpakushitsu Kakusan Koso. 2004. PMID: 15346892 Review. Japanese. No abstract available. - Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina.
Swaroop A, Kim D, Forrest D. Swaroop A, et al. Nat Rev Neurosci. 2010 Aug;11(8):563-76. doi: 10.1038/nrn2880. Nat Rev Neurosci. 2010. PMID: 20648062 Free PMC article. Review.
Cited by
- Pushing the envelope of retinal ganglion cell genesis: context dependent function of Math5 (Atoh7).
Prasov L, Glaser T. Prasov L, et al. Dev Biol. 2012 Aug 15;368(2):214-30. doi: 10.1016/j.ydbio.2012.05.005. Epub 2012 May 15. Dev Biol. 2012. PMID: 22609278 Free PMC article. - Information content differentiates enhancers from silencers in mouse photoreceptors.
Friedman RZ, Granas DM, Myers CA, Corbo JC, Cohen BA, White MA. Friedman RZ, et al. Elife. 2021 Sep 6;10:e67403. doi: 10.7554/eLife.67403. Elife. 2021. PMID: 34486522 Free PMC article. - Reduction of Glut1 in the Neural Retina But Not the RPE Alleviates Polyol Accumulation and Normalizes Early Characteristics of Diabetic Retinopathy.
Holoman NC, Aiello JJ, Trobenter TD, Tarchick MJ, Kozlowski MR, Makowski ER, De Vivo DC, Singh C, Sears JE, Samuels IS. Holoman NC, et al. J Neurosci. 2021 Apr 7;41(14):3275-3299. doi: 10.1523/JNEUROSCI.2010-20.2021. Epub 2021 Feb 23. J Neurosci. 2021. PMID: 33622781 Free PMC article. - The domestic cat as a large animal model for characterization of disease and therapeutic intervention in hereditary retinal blindness.
Narfström K, Holland Deckman K, Menotti-Raymond M. Narfström K, et al. J Ophthalmol. 2011;2011:906943. doi: 10.1155/2011/906943. Epub 2011 Apr 14. J Ophthalmol. 2011. PMID: 21584261 Free PMC article. - Regulation of Noncoding Transcriptome in Developing Photoreceptors by Rod Differentiation Factor NRL.
Zelinger L, Karakülah G, Chaitankar V, Kim JW, Yang HJ, Brooks MJ, Swaroop A. Zelinger L, et al. Invest Ophthalmol Vis Sci. 2017 Sep 1;58(11):4422-4435. doi: 10.1167/iovs.17-21805. Invest Ophthalmol Vis Sci. 2017. PMID: 28863214 Free PMC article.
References
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. - PubMed
- Acharya HR, Dooley CM, Thoreson WB, Ahmad I. cDNA cloning and expression analysis of NeuroD mRNA in human retina. Biochem Biophys Res Commun. 1997;233:459–463. - PubMed
- Akagi T, Mandai M, Ooto S, Hirami Y, Osakada F, Kageyama R, Yoshimura N, Takahashi M. Otx2 homeobox gene induces photoreceptor-specific phenotypes in cells derived from adult iris and ciliary tissue. Invest Ophthalmol Vis Sci. 2004;45:4570–4575. - PubMed
- Akagi T, Akita J, Haruta M, Suzuki T, Honda Y, Inoue T, Yoshiura S, Kageyama R, Yatsu T, Yamada M, Takahashi M. Iris-derived cells from adult rodents and primates adopt photoreceptor-specific phenotypes. Invest Ophthalmol Vis Sci. 2005;46:3411–3419. - PubMed
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, Danciger M, Davisson MT, Farber DB. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci U S A. 2000;97:5551–5556. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources