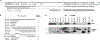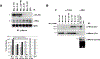Interaction of MLL amino terminal sequences with menin is required for transformation - PubMed (original) (raw)
Interaction of MLL amino terminal sequences with menin is required for transformation
Corrado Caslini et al. Cancer Res. 2007.
Abstract
Rearrangements of the mixed lineage leukemia gene MLL are associated with aggressive lymphoid and myeloid leukemias. The resulting MLL fusion proteins enforce high-level expression of HOX genes and the HOX cofactor MEIS1, which is pivotal for leukemogenesis. Both wild-type MLL and MLL fusion proteins interact with the tumor suppressor menin and with the Hoxa9 locus in vivo. Here, we show that MLL sequences between amino acids 5 and 44 are required for interaction with menin and for the transformation of hematopoietic progenitors. Blocking the MLL-menin interaction by the expression of a dominant negative inhibitor composed of amino terminal MLL sequences down-regulates Meis1 expression and inhibits cell proliferation, suggesting that targeting this interaction may be an effective therapeutic strategy for leukemias with MLL rearrangements.
Figures
Figure 1.
Menin interacts with evolutionarily conserved sequences in the NH2 terminus of MLL. A, sequence alignment of the first 167 amino acids of human MLL with Fugu MLL. Dashes were introduced to achieve better alignment. “+”, similarity. B, schematic of the MLL NH2-terminal deletions and whether they coimmunoprecipitate with menin. C, 293 cells were transfected with individual FLAG-MLL deletion fragments, immunoprecipitated with M2 anti-FLAG antibody, and probed with anti-menin antibody. The extract was also probed by anti-FLAG antibody to show the expression and expected size of the FLAG-MLL fusion proteins (bottom).
Figure 2.
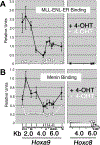
MLL fusion protein activation results in increased localization of menin at the Hoxa9 locus in MLL-ENL-ER–transformed cells. A, ChIP was done using an antibody directed against the estrogen receptor in MLL-ENL-ER and a series of PCR primers representing the Hoxa9 and Hoxc8 loci as described in Materials and Methods. The plot in (A), which is included for comparison with (B), is based on previously published data (16). Bottom, a schematic of the two Hox loci. In the presence of 4-OHT, the MLL fusion protein binds across a broad region of the Hoxa9 locus (black line). In the absence of 4-OHT (white line), MLL-ENL-ER binding is abolished and Hoxa9 expression is reduced (3). The Hoxc8 locus is not expressed in these cells and represents background levels of binding. B, ChIP using antibodies directed against menin shows that menin binding is highly correlated with binding of the MLL fusion protein. Menin binds across a broad region of the Hoxa9 locus, but not the Hoxc8 locus, in the presence of 4-OHT (black line). In the absence of 4-OHT, menin binding drops dramatically, although some residual binding remains (white line).
Figure 3.
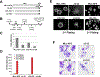
MLL sequences that interact with menin are required for transformation of hematopoietic progenitors. A, schematic of MLL deletion mutants made in the context of MLL-AF9. Left, the amino acid numbers (deletion mutants are not drawn to scale). B, schematic of the experimental procedure (see Materials and Methods). C, relative expression levels of transduced MLL-AF9 and MLL-AF9 deletion alleles analyzed by real-time quantitative RT-PCR normalized to Gapdh at 7 d of culturing in the presence of G418. D, the results of myeloid progenitor immortalization assays are summarized as the average of CFU per 2 × 104 cells plated in triplicate for each round of plating in methylcellulose. Columns, means of three independent experiments; bars, SD. E, morphology of the colonies formed after the second (left) and third (right) rounds of plating in methylcellulose of BM-transduced cells. F, Wright-Giemsa–stained cytospins of liquid cultures obtained from colonies of third-round plating in methylcellulose.
Figure 4.
Competition of MLL deletion fragments for menin binding with endogenous MLL. A, equal amounts of transfected cell extracts from GFP-positive–sorted cells were immunoprecipitated with M2 (anti-FLAG) or MLL612 (anti-MLL) antibodies, and then probed with anti-menin antibody. The representative Western blot was quantitatively determined by the ImageJ program (bottom), and the results presented as a percentage of endogenous MLL-bound menin relative to MSCV-GFP (gray columns) and as a percentage of MLL deletion mutant–bound menin relative to MSCV-MLL2–167 (black columns). B, MLL deletion mutants binding with cotransfected hemagglutinin-tagged MLL-FKBP into 293 cells. After transfections, cell extracts were immunoprecipitated with anti-FLAG and anti-MLL antibodies and Western blot analyzed with anti-hemagglutinin and anti-menin. MLL deletion mutants and MLL-FKBP coimmunoprecipitate through their association with menin. Immunoprecipitations for each MLL fragment were done at least twice (some immunoprecipitations were shown twice for figure consistency).
Figure 5.
Transduction of MLL fragments with high menin-binding avidity inhibits cell growth. A, schematic of the experimental procedure (see Materials and Methods). B, immunoprecipitation analysis of MLL deletion mutants at 2 and 8 d after GFP sorting. Equal amounts of cell extracts from GFP-positive–sorted BM cells were immunoprecipitated with M2 anti-FLAG monoclonal antibody, and then immunoblotted with anti-FLAG polyclonal antibody. At 8 d postsorting, the expression levels of MLL2–167 were significantly reduced compared with other MLL dominant negative mutants. C, GC analysis on GFP-sorted cells shows that all MLL dominant negative cells profoundly inhibited cell growth, whereas all the other transduced cells proliferated rapidly. Points, means; bars, SD. D, results of methylcellulose colony-forming assay summarized as a percentage of GFP-positive CFUs obtained from 2 × 104 transduced cells 4 d after plating. MLL-AF9–transformed cells transduced with MLL2–167, MLL2–62, or MLL Δ35–103 fragments with high menin-binding avidity show a highly reduced percentage of GFP-positive colonies, whereas for MLL2–44 dominant negative cells, the reduction was ~50% compared with MLL fragments with weak or no interaction with menin. A representative of three independent experiments. Bars, SD. E, flow cytometric analysis of GFP signal of MLL-AF9–transformed cells after MLL fragment transduction and GFP sorting. The percentage of GFP-positive cells at 4 and 11 d after cell sorting. Expression of GFP in MLL2–167, MLL2–62, or MLL Δ35–103 is rapidly down-regulated or outcompeted by GFP-negative cells. In contrast, equally high and stable percentages of GFP-positive cells are seen within all the remaining transductions.
Figure 6.
MLL dominant negative mutants interfere with Meis1 gene expression in MLL-AF9–transformed BM cells. A, schematic of the experimental procedure. MLL-AF9–transformed BM cells obtained from the third round of passage in methylcellulose were spin-infected (SI) with MSCV-GFP empty vector or encoding for the MLL deletion mutants indicated. The GFP-positive cells were flow-sorted at 2 and 7 d following transductions and total RNA was extracted 24 h later. B, quantitative real-time PCR comparison of MLL-AF9, Meis1, and HoxA9 expression among MLL-AF9 immortalized BM cells at 9 d posttransduction and MLL-AF9–transformed cells at the third round of passage in methylcellulose. C, quantitative real-time PCR for Meis1 and Hoxa9 expression levels normalized to Gapdh at 3 d posttransduction. D, quantitative real-time PCR for Meis1 and Hoxa9 expression levels normalized to Gapdh at 8 d posttransduction. Columns, percentages relative to MLL-AF9–transformed/MSCV-GFP vector–transduced BM cells; bars, SD.
Figure 7.
MLL dominant negative mutants inhibit the colony-forming ability of wild-type myeloid progenitors. A, experimental procedure (see Materials and Methods). B, interaction of transduced MLL dominant negative mutants with endogenous menin in GFP-sorted BM cells. Equal amounts of cell extracts from GFP-positive–sorted BM cells were immunoprecipitated with M2 anti-FLAG monoclonal antibody and then immunoblotted with anti-menin polyclonal antibodies. A 5% aliquot of each cell lysate was analyzed by Western blot as control for endogenous levels of menin (Input). C, results of methylcellulose colony-forming assay summarized as a percentage of GFP-positive CFU obtained from 2 × 104 transduced cells at 7 to 10 d from plating. Wild-type myeloid progenitor cells transduced with MLL dominant negative fragments with high menin-binding avidity show a reduced percentage of GFP-positive colonies compared with cells transduced with MLL fragments with weak or no interaction with menin (MLL2–35) or GFP alone. Bars, SD. D, morphology and density of GFP-positive colonies at 14 d from second methylcellulose plating. E, Wright-Giemsa–stained cytospins of cells obtained from colonies at the second round of plating in methylcellulose.
Similar articles
- PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute Myeloid Leukemias Characterized by a Core Transcriptome of the MLL-Rearranged Disease.
Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun AG, Zuo Z, Gurbuxani S, Arnovitz S, Weng H, Wang Y, Li S, Huang H, Neilly MB, Wang GG, Jiang X, Liu PP, Jin J, Chen J. Li Z, et al. Cancer Res. 2016 Feb 1;76(3):619-29. doi: 10.1158/0008-5472.CAN-15-1566. Epub 2016 Jan 8. Cancer Res. 2016. PMID: 26747896 Free PMC article. - The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis.
Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. Yokoyama A, et al. Cell. 2005 Oct 21;123(2):207-18. doi: 10.1016/j.cell.2005.09.025. Cell. 2005. PMID: 16239140 - TET1 plays an essential oncogenic role in MLL-rearranged leukemia.
Huang H, Jiang X, Li Z, Li Y, Song CX, He C, Sun M, Chen P, Gurbuxani S, Wang J, Hong GM, Elkahloun AG, Arnovitz S, Wang J, Szulwach K, Lin L, Street C, Wunderlich M, Dawlaty M, Neilly MB, Jaenisch R, Yang FC, Mulloy JC, Jin P, Liu PP, Rowley JD, Xu M, He C, Chen J. Huang H, et al. Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):11994-9. doi: 10.1073/pnas.1310656110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818607 Free PMC article. - Deregulation of the HOXA9/MEIS1 axis in acute leukemia.
Collins CT, Hess JL. Collins CT, et al. Curr Opin Hematol. 2016 Jul;23(4):354-61. doi: 10.1097/MOH.0000000000000245. Curr Opin Hematol. 2016. PMID: 27258906 Free PMC article. Review. - Menin as a hub controlling mixed lineage leukemia.
Thiel AT, Huang J, Lei M, Hua X. Thiel AT, et al. Bioessays. 2012 Sep;34(9):771-80. doi: 10.1002/bies.201200007. Epub 2012 Jul 24. Bioessays. 2012. PMID: 22829075 Free PMC article. Review.
Cited by
- Distinct pathways regulated by menin and by MLL1 in hematopoietic stem cells and developing B cells.
Li BE, Gan T, Meyerson M, Rabbitts TH, Ernst P. Li BE, et al. Blood. 2013 Sep 19;122(12):2039-46. doi: 10.1182/blood-2013-03-486647. Epub 2013 Aug 1. Blood. 2013. PMID: 23908472 Free PMC article. - SUV39H1 regulates the progression of MLL-AF9-induced acute myeloid leukemia.
Chu Y, Chen Y, Guo H, Li M, Wang B, Shi D, Cheng X, Guan J, Wang X, Xue C, Cheng T, Shi J, Yuan W. Chu Y, et al. Oncogene. 2020 Dec;39(50):7239-7252. doi: 10.1038/s41388-020-01495-6. Epub 2020 Oct 9. Oncogene. 2020. PMID: 33037410 Free PMC article. - A novel Menin-MLL1 inhibitor, DS-1594a, prevents the progression of acute leukemia with rearranged MLL1 or mutated NPM1.
Numata M, Haginoya N, Shiroishi M, Hirata T, Sato-Otsubo A, Yoshikawa K, Takata Y, Nagase R, Kashimoto Y, Suzuki M, Schulte N, Polier G, Kurimoto A, Tomoe Y, Toyota A, Yoneyama T, Imai E, Watanabe K, Hamada T, Kanada R, Watanabe J, Kagoshima Y, Tokumaru E, Murata K, Baba T, Shinozaki T, Ohtsuka M, Goto K, Karibe T, Deguchi T, Gocho Y, Yoshida M, Tomizawa D, Kato M, Tsutsumi S, Kitagawa M, Abe Y. Numata M, et al. Cancer Cell Int. 2023 Feb 25;23(1):36. doi: 10.1186/s12935-023-02877-y. Cancer Cell Int. 2023. PMID: 36841758 Free PMC article. - Menin represses tumorigenesis via repressing cell proliferation.
Wu T, Hua X. Wu T, et al. Am J Cancer Res. 2011;1(6):726-39. Epub 2011 May 16. Am J Cancer Res. 2011. PMID: 22016823 Free PMC article. - The role of HTS in drug discovery at the University of Michigan.
Larsen MJ, Larsen SD, Fribley A, Grembecka J, Homan K, Mapp A, Haak A, Nikolovska-Coleska Z, Stuckey JA, Sun D, Sherman DH. Larsen MJ, et al. Comb Chem High Throughput Screen. 2014 Mar;17(3):210-30. doi: 10.2174/1386207317666140109121546. Comb Chem High Throughput Screen. 2014. PMID: 24409957 Free PMC article. Review.
References
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med 2004;10:500–7. - PubMed
- Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 2002;10:1107–17. - PubMed
- Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 2002;10:1119–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous